Research Article - (2025) Volume 33, Issue 2
Received: 20-May-2024, Manuscript No. IPQPC-24-19976; Editor assigned: 22-May-2024, Pre QC No. IPQPC-24-19976 (PQ); Reviewed: 02-Jun-2024, QC No. IPQPC-24-19976; Revised: 16-Apr-2025, Manuscript No. IPQPC-24-19976 (R); Published: 23-Apr-2025, DOI: 10.36648/1479-1064.33.2.51
Background: The abnormal regulation of circular RNA (circRNA) levels is commonly identified in human diseases, particularly malignant tumors. Recently, the diagnostic value of circRNAs has received increased attention. The detailed mechanisms of various cancer and circular RNAs need more research to clarify, including hepatocellular carcinoma HCC.
Methods: We utilized quantitative Real-Time fluorescence Polymerase Chain Reaction (RT-qPCR) to measure the expression level of hsa_circ_0000038 in paired Hepatocellular Carcinoma (HCC) and adjacent noncancerous liver tissues. GO annotation and enrichment analysis were used to examine the potential downstream pathways. RT-qPCR and Western blotting were conducted to evaluate the expression of thep53/p21pathway. CCK-8, wound closure and Tran’s well assays were used to measure cell proliferation, migration and invasion. Luciferase and chromatin immune precipitation assays were used to investigate the interactions between miR-92a-2-5p and hsa_circ_0000038.
Results: Levels of hsa_circ_0000038 were down regulated in HCC tissues and cells. Overexpression ofhsa_circ_0000038 inhibited tumor growth in vivo and blocked the hepatocarcinoma cell cycle at the G0–G1 phase and repressed cell proliferation, invasion and migration of HCC cells in vitro, while cotransfection of miR-92a-2-5p partially attenuated the effects mediated by hsa_circ_0000038. The expression of miR-92a-2-5p was decreased in HCC tissues and promoted cell proliferation and the cell cycle in vitro. hsa_circ_0000038 acted as a sponge for miR-92a-2-5p and Tp53 gene was the target ofmiR-92a-2-5p. Hsa_circ_0000038 inhibited the progression of tumor growth by inhibiting the miR-92a-2-5p/p53/p21 axis.
Conclusion: Our study reveals aberrant circRNA expression profiles in HCC tissues. Hsa_circ_0000038 regulates themiR-92a-2-5p/p53/p21 axis and be involved in HCC development.
Circular RNA (circRNA); Hepatocellular Carcinoma (HCC); miR-92a-2-5p; p53/p21 pathway; Tumor progression
Hepatocellular Carcinoma (HCC) is the most prevalent form of primary liver cancer. Studies have estimated that chronic HBV/HCV infection accounts for 80% of HCC cases. According to the latest update, the annual incidence of HCC is 1%–6% and HCC is the third highest cause of cancer-related deaths globally. More than 905,677 cases were diagnosed and 830,180 HCC patients died in 2020. The rate of new HCC cases has more than doubled in the past three decades in the West, leading to an increased public health burden [1,2]. However, the current diagnostic strategies for HCC are classified as invasive or non-invasive (e.g., the liver imaging reporting and data system) methods. Both methods frequently apply to the middle-late phase of HCC [3,4]. In addition, the sensitivity and specificity of current tumor biomarkers are unsatisfactory. Despite the advancements in treatment strategies (such as catheter-based loco-regional treatment, curative resection, immune checkpoint inhibitors, local ablation and chemotherapy), the prognosis for patients with HCC remains very poor due to tumor metastasis and recurrence, adverse side effects and drug resistance [5-8]. Therefore, there is an urgent need for new and effective molecular biomarkers. Circular RNAs (circRNAs) lack 5'-3' polarity and polyadenine tails. They are transcribed by RNApolymerase II with the same transcriptional efficiency as linear RNA. These properties indicate that circRNAs are more resistant to RNase than linear RNA. Aberrant expression of circRNAs is a crucial factor in a variety of malignancies, including HCC [9]. The impact of circRNAs on tumors is twofold: They function as both a tumor suppressor and a proto-oncogene to regulate tumor growth and progression. MicroRNAs (miRNAs) are a class of small non-coding RNAs ranging from 17 to 25 nt in length. MicroRNAs regulate gene expression by binding to the miRNA response elements in the coding domain sequence or 3’ Untranslated Region (UTR) of target mRNAs, thereby down regulating gene expression. Studies of how circRNAs modulate mRNA expression mainly focus on the ceRNA mechanism. ceRNAsare a class of transcripts that can competitively regulate miRNAs at the post-transcriptional level and can participate in a wide range of biological processes through the circRNA/miRNA/mRNA axis. Mounting evidence indicates that circRNAs are involved in the development of HCC via sponging miRNAsto alter the activity of signaling pathways and host genes [10,11]. For example, circMAPK9 spongesmiR642 and affects the progression of HCC. Circ-ZEB1.33, hsa_circ_0001459, and circ_0011232 have been reported to be functional miRNA sponges in HCC [12- 15]. Furthermore, MiR-92a is a widely studied microRNA, in particular, its role in inflammatory and cancer diseases. Several studies have shown thatmiR-92 promote the expression of numerous cancer genes and is involved in multiple classical cancer-associated pathways [16–20]. MiR-92a promotes hepatocellular carcinoma cell proliferation and invasion by Recombinant Forehead Box Protein A2 (FOXA2) targeting. MiR92a-3p regulates the phosphatase and tensin homolog deleted on chromosome ten/ protein kinase B (PTEN/AKT) pathway in HCC while miR-92a-2-5p increases liver cancer cell invasion by altering Androgen Receptor (AR)/PH Domain Leucine-rich Repeat Protein Phosphatase (PHLPP)/p-AKT/β-catenin signaling [19]. Tp53 (Tumor protein 53(Tp53)) is susceptible to inactivation in hepatocellular carcinoma, suggesting thatp53 plays a key role in the development and progression of HCC. Studies have shown that p53alterations are associated with tumor differentiation, vascular invasion and tumor staging in HCC. p53 and p21 are key regulators of cell cycle arrest. It has been shown that p53 induces cell cycle arrest and apoptosis at the G1/S border mainly through direct transcriptional induction of CDK inhibitors p21and puma/noxa. In addition, the activation of p53 can also be facilitated by circRNAs. circRNAs showed a differential expression in various cancers when p53 is activated. Therefore, understanding how circRNAs target miRNAs to regulate p53 is beneficial for a deeper exploration of howp53 is activated to exert anti-tumor effects in HCC. Here, we discovered, for the first time, that hsa_circ_0000038 as a regulator of the miR92a-2-5p, which in turn modulates the p53/p21 proteins to inhibit tumor progression in HCC. These findings contribute to the elucidation of a novel molecular mechanism involving circRNA and expand our comprehension of the p53 pathway and competing endogenous RNA (ceRNA) in HCC.
Clinical Specimens and HCC Tissue Collection
The research was approved by the Ethics Committee of the Affiliated Hospital of Yunnan University, Kunming, China and all subjects signed written informed consent forms. The inclusion criteria for patients are as follows: (1) clinically diagnosed as primary liver cancer; (2) no prior treatment with other antitumor drugs, radiotherapy or chemotherapy. Exclusion criteria were: (1) combined chronic hepatitis virus infection; (2) those who have or have had cirrhosis de compensation; (3) patients with serious organ lesions and cardiopulmonary insufficiency before hospital; (4) co-infection with HIV; (5) progression to chronic hepatitis B-associated liver cancer or other tumors; (6) patients with severe anemia normal nutrition; (7) abnormal platelet and coagulation function. Fresh blood and tissue samples (20 pairs of HCC tissues and adjacent non-tumor tissues) were collected from patients. Blood samples were centrifuged at 3300 rpm for 5 min to obtain serum samples. All samples were frozen in liquid nitrogen and kept at −80°C until use.
Cell Culture
The human HCC cell lines (Huh7, MHCC-97H and HepG2, SK hep1 cells) were purchased from the Chinese Academy of Sciences (Kunming Institute of Zoology, CAS, China) and the human hepatic progenitor cell line (HepaRG) was donated by the Department of Hepatobiliary Surgery, Second Affiliated Hospital of Kunming Medical University, Kunming, China. Cells were cultured in DMEM (Gibco, GrandIsland, NY, USA) containing 10% inactivated fetal bovine serum (FBS, Gibco), 100 U/mL streptomycin and100 U/mL penicillin in a moist incubator at 37°C and 5% CO2. Assays were performed during the logarithmic growth phase of the cells.
Plasmid Construction and Cell Transfection
The whole-length sequence of hsa_circ_0000038 was amplified and chemo-synthesized by PCR. The company gene seed (Guangzhou, China) inserted these segments into the pLC5-ciR vector. Artificial ranking sequences and splice acceptor/donor sequences are both present in the pLC5-ciR vector. AnsiRNA for hsa_circ_0000038 knockdown was constructed to target its back-splice sequence by Gene Pharma (Suzhou, China). When 50–60% confluent, cells were transfected with plasmids and siRNAs using Lipofectamine™ 3000 Transfection Reagent (In vitro gen, Shanghai, China). The sequence of siRNAis.
RNase R Assay
Total RNA was extracted from HepG2 cells described above. The RNA was divided into two parts; one was treated with RNase R (RNase R+) and the other was not treated (RNase R-). The reaction condition was 37°C for 20 min. Finally, the RNA was purified and recovered for agar-gel electrophoresis.
RNA Isolation and Quantitative Real-Time PCR (RT qPCR)
TRIzol reagent (Invitrogen, USA) was used to extract total RNA from cells, tissues and plasma samples. After measuring the RNA concentration, RNA was stored at −80°C for future analysis. Real-time PCRusing and ABI 12k Fast Real-Time PCR System (Applied Biosystems, USA) was conducted using Fast Start Universal SYBR Green Master (ROX) (Roche, Switzerland) and three replicate wells for each sample. The relative expression was calculated with the 2 − ΔΔCt method. The PCR was as follows: Initial denaturation for 5 min at 95°C followed by 40 cycles at 95°C for 15 s and 1 min of annealing/extension at 60°C. β-actin was used to normalize the mRNA expression levels.
Actinomycin D Treatment
The cells were cultured and divided into two groups according to the experimental design. HepG2 cells in each group were treated with actinomycin D (2 µg/ml) for 0, 12 and 24 h. At each time point, cells were collected and the total RNA was extracted. Genomic DNA was removed using the PrimeScript RT Reagent Kit with gDNA eraser (Takara, Tokoyo, Japan) and cDNA was synthesized by reverse transcription. RT-qPCR was performed using FastStart Universal SYBR Green Master (ROX) to detect the levels of targetRNA and reference RNA.
Cell Proliferation Assay
Cell standard curves were made using a CCK8 assay kit (Invitrogen, USA) the manufacturer’s protocol and then the appropriate number of cells to be inoculated and the measurement time were determined. Based on the specific transfection, cell suspensions (8 × 103 cells/well) were inoculated into 96-well plates and pre-cultured in the incubator for 24 hours. Before each test, 10 µl CCK8 solutions was added to each well and incubated for 2.5 hours protected from light. The absorbance was measured at 450 and 650 nm using a Varioskan Flash system (Thermo, USA). Measurements were made at 0, 24, 48 and 72 hours after the cells had attached.
Wound-Healing Assay
An Ibidi cell insert (Ibidi, Germany) was applied to make a scratch in the middle of the well. After adding70 µl (1 × 105 cells/ml) of cell suspension during the logarithmic growth phase to each chamber of the insert, the cells were cultured for 24 hours in serum-free DMEM. The insert was then removed with forceps and the wells were washed gently with cell-free medium or PBS. Then, 2 ml of the FBS-free medium was added to the wells for incubation. Images were taken using an inverted microscope at 0 and 24 hours and the cell coverage of the scratch area was measured.
In vitro Invasion Assay
Matrigel matrix (Corning, USA) was mixed 1:4 with FBS-free medium at 4°C. The above mixture was coated onto the upper chamber of the trans well insert. The 24-well plates were incubated for 2 hours at 37°C to coagulate the gel and then 500 µl DMEM with 5% FBS was added to the lower chamber. The upper chamber was added to the wells of the 24-well plate. Transfected cells in the logarithmic growth phase were rinsed with PBS and suspended in serum-free DMEM. The upper concentration was adjusted to 1 × 105/ml and 150 µl were added to the upper chamber. After incubating for 48 hours, the chamber was permeabilized by 4% paraformaldehyde for 20-30 min and then stained with 0.5% crystal violet for 10 min. The invaded cells were counted under a light microscope in at least five random fields.
Western Blotting
We used RIPA buffer to extract protein from transfected cells and determined the protein concentration using a BCA kit (Beyotime, Shanghai, China). Forty nano grams of protein samples were separated by electrophoresis on a 10% polyacrylamide gel and then transferred to PVDF membranes (Millipore, Billerica, MA, USA). The membrane was placed in blocking solution (Millipore) for 4 hours at room temperature and then incubated with primary antibody against PHACTR4 (1:5000, Proteintech, Beijing, China), p53 (1:1000, Proteintech), p21 (1:1000, Proteintech) and β-actin (1:2000, Proteintech) in a shaker at 4°C overnight. A horseradish peroxidase-conjugated secondary antibody was added to the membranes. Images were captured by ChemiDoc XRS+ (Bio rad, USA) and the protein expression level was calculated using ImageJ software.
Dual-Luciferase Reporter Assay
According to star base v2.0 prediction, the predicted binding site of hsa_circ_0000038 to mir-92a-2-5p was mutated by segmentation. Mir-92a-2-5p may bind to hsa_circ_0000038 at two separate sites and so we constructed two mutants of hsa_circ_0000038 (mut1 and mut5). The wild-type and mutanthsa_circ_0000038 or wild-type and mutant Tp53 3'UTRs were inserted into the pmirGLO dual-luciferase vector. 5 × 104 cells/ml HEK-293T cells in 96-well plates were co transfected with the vectors (miR92a-2-5p mimics/NC and hsa_circ_0000038 wild/mut, miR92a-2-5pmimics/NC and Tp53 wild/mut). The cells were quantified using a dual luciferase reporter analysis kit (Beyotime). Following transfection for 24 hours, the luciferase signals were read using a dual-luciferase reporter assay system (Promega, USA).
RNA Immunoprecipitation Assay
The RNA Immunoprecipitation (RIP) assay was performed using an RNA immunoprecipitation kit (Geneseed, China) with AGO2-specific antibodies (Cat No. 67934-1-Ig, Proteintech) following the manufacturer’s instructions. The immuno precipitated RNAs were detected by RT-qPCR to measure the level of hsa_circ_0000038 (or Tp53). A mouse isotype antibody (IgG) was applied as the control.
Colony Formation Assay
Transfected HCC cells were plated into six-well plates and incubated under 5% CO2 at 37°C. After 2 weeks, the cells were washed with Phosphate-Buffered Saline (PBS) and fixed with 4% paraformaldehyde for 20 min. Crystal violet was used to stain the cells. Each colony containing at least 50 cells under a microscope was counted as valid.
Xenografts in Nude Mice
The Second People's Hospital of Yunnan Province’s ethics committee approved the mice xenograft studies. Twenty male (n=5/group) BALB/c-nu/nu mice were acquired from Kunming Medical University and were kept in a pathogen-free isolator. Feeding and management were carried out under aseptic conditions. Huh 7 cells stably transfected with pLC5-ciR or hsa_circ_0000038 were combined with 10% stromal gel (Corning Costar, USA) at a cell concentration of 5 ×106 cells/ml and subcutaneously injected into the axillae of mice. Calipers were used to quantify tumor dimensions (mm) 7 days after transplantation. The diameter of the xenograft tumors and the body weight were measured daily. Three weeks after the tumors were implanted, the mice were sacrificed. The tumor weight (mg) was measured using an electronic balance. The tumor volume (mm3) was determined as follows: Volume=0.5 × diameter3
CircRNA/miRNA Interaction Annotation and Gene Ontology (GO) Enrichment Analyses
miRanda and RNA hybrid Databases were used to determine miRNAs and circRNAs that may interact. The R package cluster Profiler was used to conduct GO enrichment analysis of the host genes to further predict the signaling pathways of circRNAs.
Statistical Analyses
All data statistics were performed using SPSS17.0 for Windows and were plotted using GraphPad Prism9.0. The expression level of hsa_circ_0000038 was analyzed using a paired t-test to compare between carcinomatous and normal tissues. The clinical and pathological characteristics of hsa_circ_0000038expression in HCC patients were assessed by a two-sample t-test. The mean ± SD values are shown. p<0.05 was considered statistically significant.
The Expression Levels of hsa_circ_0000038 are Down Regulated in Tissues from Liver Cancer Patients and Hepatoma Cell Lines
Our research on the hepatitis B virus found that hsa_circ_0000038 suppressed the proliferation of HepG2.2.15 cells (Figure 1A). Therefore, this prompted us to consider whether this factor is associated with liver tumor disease. Twenty pairs of HCC patients (n=20) were recruited. The expression levels of hsa_circ_0000038 were determined in liver cancer tissues and their para-cancerous tissue specimens using RT-qPCR. The expression was significantly down regulated in liver cancer tissues compared with their paired para-cancerous tissues (Figure 1B; p<0.01). In parallel, we measured the expression levels of the circ RNA-derived host gene Phactr4 and found no significant difference (p>0.05; Figure 1C). Four commonly used hepatocellular carcinoma cell lines (Huh7, SK-hep1, HepG2 and MHCC97-H) and hepatic progenitor cells (HepaRG) were selected. The expression levels of hsa_circ_0000038 were significantly lower in hepatocellular carcinoma cells compared with HepaRG (Figure 1D). The correspondence between the translation level and transcription level was verified. The PHACTR4 protein was down-regulated inhuman liver cancer tissues and in HCC cells (Figure 1).
Figure 1: Characterization of hsa_circ_0000038 in liver cancer tissues and cells.
Note: (A) The proliferative abilities of Hep G2 cells between the hsa_circ_0000038-transfected group and the vector transfected group were measured by the CCK-8 assay. (B) Expression levels of hsa_circ_0000038 in HCC tissues in comparison with matched normal tissues were measured using RT-qPCR. (C) Expression levels of Phactr4 in HCC tissues in comparison with matched normal tissues were measured using RT-qPCR. (D) RT-qPCR verified the basal expression level of hsa_circ_0000038 in different five liver cell lines. (E) Western blot analysis of PHACTR4 protein in HCC tissues and normal tissues, β-actin was used as a loading control. (F) Western blot analysis of PHACTR4 protein in HepaRG and the other four liver cancer cell lines. *p<0.05, **p<0.01,***p< 0.001.
Hsa_circ_0000038 Inhibits Proliferation, Migration and Invasion of HCC Cells
The loss-of-function assay was conducted in HCC cells. The basal expression level of hsa_circ_0000038was initially determined in four hepatocellular carcinoma cell lines. We chose Huh7 and SK-hep1 cells to over express the hsa_circ_0000038; HepG2 and MHCC97-H cells were chosen to silencehsa_circ_0000038. Due to their limited invasion ability, the invasion assay was performed using HepG2, SK Hep1 and MHCC97-H cells. After exogenous transfection of the hsa_circ_0000038 plasmid, the expression of hsa_circ_0000038 was elevated in Huh7 and SK-Hep1 cells compared with the circ-NC, whereas after transfection of si circ_0000038 in HepG2 and MHCC97-H cells, the expression ofsi_circ_0000038 (A siRNA that knocks down hsa_circ_0000038) was specifically silenced compared with si circNC (NC control to siRNA) (Figure 2A and Figure 2B). At the same time, overexpression or silencing ofhsa_circ_0000038 failed to upregulate or knock down the expression of the host gene PHACTR4 (Figure 2C). The CCK-8 assay detected that the up regulation of hsa_circ_0000038 resulted in a decrease in cell proliferation in Huh7 and SK-Hep1 cells, as indicated by lower OD values within 72 hours, compared with the circ-NC group (Figure 2D and 2E). In the cell cycle assays, the overexpression of hsa_circ_0000038 in Huh7 and SK-hep1 cells led to an increase in the number of cells transitioning from S and G2/M phases to G0/G1 phases, compared with the circ NC-transfected group. When si-circ_0000038 was transfected into HepG2 and MHCC97-H cells, it led to a higher transformation of cells from G0/G1 phases to S and G2/M phases (Figure 2F and 2G). A scratch experiment was performed to evaluate cell migration and Transwell assays were used to assess invasion. In comparison to si-NC treatment, the number of migrating and invaded cells increased following the administration of si-circ_0000038 (Figure 2H and 2I). Conversely, the overexpression of hsa_circ_0000038 decreased the number of migrating and invading cells compared with the circ-NC-transfected group (Figure 2J and 2K). The behavior of the cells was observed by colony formation assay. Cell density and cell clonality was significantly higher in Huh7 cells transfected with hsa_circ_0000038 than in Huh7 cells transfected with circ-NC.
Figure 2: hsa_circ_0000038 decreased cell proliferation, migration and invasion and changed the polarization of the cell cycle in vitro.
Note: Verification of hsa_circ_0000038 knock down by RT qPCR analysis. (A,B) Verification of hsa_circ_0000038 knockdown or over expression by RT-qPCR analysis. (C) Transfection of hsa_circ_0000038 or knockdown of hsa_circ_0000038 did not affect the expression level of Phactr4. (D,E) Huh7 and SK-hep1 cells were transfected with hsa_circ_0000038. HepG2 and MHCC97-H cells were transfected with si-hsa_circ_0000038. Cell growth was measured by CCK-8 assay. Cell proliferation was repressed by hsa_circ_0000038 but promoted by si-hsa_circ_0000038. (F,G) The down regulation or over expression of hsa_circ_0000038 changed the cell cycle. hsa_circ_0000038 could arrest the cell cycle in the G0/G1 phase. Knockdown of hsa_circ_0000038 promoted the cell cycle change toward the S-G2/Mphase. (H) hsa_circ_0000038 over expression suppressed the migration of SK-hep1 cells. (I) hsa_circ_0000038 down regulation promoted the migration capacities of HepG2 cells. (J) hsa_circ_0000038 overexpression suppressed the invasion of Huh7 and SK-hep1, HepG2 cells. (K) hsa_circ_0000038 down regulation promoted the invasion of HepG2 and MHCC97-H cells. *p<0.05, **p<0.01, ***p< 0.001.
Hsa_circ_0000038 has a Ring Structure and High Stability
Hsa_circ_0000038 was significantly down regulated in liver samples from patients with hepatocellular carcinoma and in hepatocellular carcinoma cell lines. Bio-informatic analyses using circBase and Circ interactome revealed thathsa_circ_0000038 is derived from exons 6 and 7 of the PHACTR4 gene. Additionally, PCR primers were separately designed around the upstream and downstream regions of the hsa_circ_0000038 target fragment. These are referred to as convergent primers and the fragment to be amplified is located between the upstream and downstream primers. The trans-loop site of hsa_circ_0000038 is shown. The genomic length of hsa_circ_0000038 was 2738 nt, while the spliced sequence length was 783 nt (Figure 3A). The body region is identical to the linear RNA, except for at the trans-loop RNA primer cut point. A pair of divergent primers was designed to specifically target the back splice junction site. Both pairs of primers were amplified simultaneously using PCR and the size of the PCR product was detected by electrophoresis to confirm the loop-forming property of hsa_circ_0000038 (Figure 3B). The circular structure of hsa_circ_0000038 was confirmed using the RNase R-digestion assay. The results revealed thathsa_circ_0000038 was more resistant to the RNase R enzyme compared with linear RNAs in HepG2 cells (Figure 3C). Similarly, actinomycin D was used to verify the stability of hsa_circ_0000038. With increasing time of actinomycin D treatment, the expression of linear RNA decreased but circRNA did not change significantly (Figure 3D). These results indicate that hsa_circRNA_0088036 has a closed-loop structure.
Hsa_circ_0000038 suppresses tumor growth in vivo: To evaluate the tumor suppressive effect of hsa_circ_0000038 in vivo, Huh7 cells stably expressing the control or hsa_circ_0003258 were intravenously injected into the tail vein of BALB/c nude mice. The weight was measured for 27 days. Mice injected with cells overexpressing hsa_circ_0000038 had less tumor tissue than mice injected with control cells (Figure 3E). There was a significant difference in weight and tumor volume between the hsa_circ_0000038 and control groups (Figure 3F and 3G). These findings suggest that overexpression of hsa_circ_0000038 prevents the proliferation of HCC in vivo.
Figure 3: Closed-loop structure and stability verification of hsa_circ_0000038 and tumour formation experiments in nude mice.
Note: (A) The genomic loci of the hsa_circ_0000038 gene. Hsa_circ_0000038 is synthesized at thePHACTR4 gene locus containing exons 6 to 7. The back-splice junction of hsa_circ_0000038 was identified by Sanger sequencing. (B) After PCR with convergent and divergent primers, gel electrophoresis was performed. The amplification lengths of different primers were as expected. In cDNA, convergent primers produced a 783-nt product, while divergent primers produced a 238-nt product. In gDNA, convergent primers produced a 2783-nt product, while divergent primers did not produce a specific band. (C) hsa_circ_0000038 was confirmed as a circRNA that is resistant to RNase R treatment compared with the linear RNA. (D) Expression trends of hsa_circ_0000038 and linear RNA in cells treated with 2 μg/mlactinomycin D for 24 h. Each experiment was performed at least three times independently. (E) Nudemice (n=5) aged 3–4 weeks were fed in a sterile environment and divided into two groups. One group was injected with Huh7 cells transfected with hsa_circ_0000038 under high pressure and the other group was injected with NC for comparison. The weight of the nude mice was recorded every three days. After 21 days, the mice were killed to remove the tumor and the tumor volume in each group was measured. (F,G) Mouse body weight and tumor volume were decreased in the overexpression hsa_circ_0000038 group, suggesting hsa_circ_0000038 could inhibit the growth of tumors. *p<0.05, ***p< 0.001.
Hsa_circ_0000038 acts as a Sponge for miR-92a-2-5p
Prediction of hsa_circ_0000038 target-bound miRNA on the websites miRanda and RNAhybrid. The websites revealed that miR-92a-2-5p be a target of hsa_circ_0000038 (Figure 4A). miR-92a-2-5p was up regulated in HCC tumor tissues compared with para-neoplastic tissues (Figure 4B). However, miRNA expression in this patient cohort did not show a negative correlation with hsa_circ_0000038, possibly due to the sample size being too small (Figure 4C). Overexpression ofhsa_circ_0000038 in Huh7 and SK-Hep1 cells led to decreased expression of miR-92a-2-5p (Figure 4D and 4E). In contrast, silencing hsa_circ_0000038 through siRNA transfection led to increased expression ofmiR-92a-2-5p (Figure 4F and 4G). The expression of miR-92a-2-5p was significantly down regulated in Huh7 and HepG2 cells overexpressing hsa_circ_0000038. Mut part1-circ_0000038 and Mut part5-circ_0000038 were identified by mutating WT circ_0000038 and miR-92a-2-5p, which were constructed with two different complementary sites (Figure 4H), to elucidate the binding site. HEK-293T cells were initially transfected with hsa_circ_0000038 WT or MUT, respectively and subsequently transfected with miR-92a-2-5p 6 hours later. The results from the dual-luciferase reporter gene assay indicated that cells transfected with wild-type-circ_0000038 had significantly lower fluorophore activity compared with cells transfected with mut-hsa_circ_0000038 due to the overexpression of miR-92a-2-5p (Figure 4I). In the RIPassay, circRNAs were found to be preferentially enriched in miRNA ribo-nucleo protein complexes containing AGO2 compared with the control IgG (Figure 4J). These results suggest that hsa_circ_0000038can regulate miR-92a-2-5p through target binding in HCC.
Figure 4: Hsa_circRNA_0000038 functions as a sponge for miR-92a-2-5p.
Note: (A) Potential binding sites of hsa_circRNA_0000038 and miR-92a-2-5p. (B) miR-92a-2-5p expression in 20 paired human HCC and adjacent normal liver tissues. (C) miR-92a-2-5p levels were measured in Huh7 cells transfected withhsa_circ_0000038. (D) miR-92a-2-5p levels were measured by RT-qPCR in SK-hep1 cells transfected withhsa_circ_0000038. miR-92a-2-5p levels were down regulated while transfected with hsa_circ_0000038. (E,F) miR-92a-2-5p levels were measured by RT-qPCR in cells transfected with si-hsa_circ_0000038. miR-92a-2-5p levels were up regulated while transfected with si-hsa_circ_0000038. (G-I) Dual luciferase reporter assays demonstrated that miR-92a-2-5p is a direct target of hsa_circRNA_0000038. This Figure (G) showed that miR-92a-2-5p and hsa_circRNA_0000038 could be predicted to directly bind to two different sites, then we designed variations at these two potential sites, the two mutant plasmids were named mut1-hsa_circ_0000038 and mut5-hsa_circ_0000038 respectively. The Figure (H) showed that compared with cells transfected with mut1 hsa_circ_0000038, cells transfected with wt1 circ_0000038cells had significantly lower fluorophore activity because of miR-92a-2-5p overexpression. The Figure (I) indicated that there are no remarkable difference between wt5-circ_0000038 and mut5- circ_0000038after miR-92a-2-5p overexpression. (J) RIP assays showed there was a direct target site between miR-92a-2-5p and hsa_circ_0000038. * p<0.05, **p<0.01, ***p< 0.001.
MiR-92a-2-5p Counteracts the Phenotypic Alterations Induced by Transfection of hsa_circ_0000038 in Hepatocellular Carcinoma Cells
Transfection of miR-92a-2-5p mimics partly counteracted the miR-92a-2-5p down regulation induced by the overexpression of hsa_circ_0000038 in Huh7 and SK-Hep1 cells. Transfection of a miR-92a-2-5p inhibitor to the si-circ38 groups of MHCC97-H or HepG2 cells could partly counteract the effect in a similar way (Figure 5A and 5B). Huh7 and SK-Hep1 cell proliferation was inhibited by hsa_circ_0000038, but the effect was lessened by the application of miR-92a-2-5p mimics (Figure 5C and 5D). The suppression of scratch migration, as well as Transwell invasion, was also partly counteracted in hsa_circ_0000038-overexpressing Huh7 and SK-Hep1 cells (Figure 5E–H). The cell cycle assay showed that the cycle of tumor cells shifts from the S and G2/M phases to G0/G1 phases and the direction of the above shift begin to reverse after transfection with miR-92a-2-5p mimics (Figure 5I and 5J). These data confirm a direct interaction between hsa_circ_0000038 and miR-92a-2-5p and suggest that miR-92a-2-5p can counter act the inhibitory effect of hsa_circ_0000038 overexpression on the development of liver malignancies.
Figure 5: Hsa_circRNA_0000038 functions could be recovered by miR-92a-2-5p mimics including cell proliferation migration and invasion and changed the polarization of the cell cycle in vitro.
Note: (A, B) miR-92a-2-5p level was measured by RT-qPCR. (A) miR-92a-2-5p expression level in Huh7 and SK-hep1 cells transfected with (hsa_circ_0000038+miR-92a-2-5p mimics)was down regulated compared with cells transfected withhsa_circ_0000038 alone. (B) miR-92a-2-5p expression levels in HepG2 and MHCC97-H cells transfectedw ith (si hsa_circ_0000038+miR-92a-2-5p inhibitor) was up regulated compared with cells transfected withsi-hsa_circ_0000038 alone. (C,D) The addition of a miR-92a-2-5p mimics partially rescued the growth ofHuh7 and SK-hep1 cells transfected with hsa_circ_0000038, as determined by the cell proliferation assay. In the contrast, the addition of a miR-92a-2-5p inhibitor partially inhibited the growth of HepG2 andMHCC97-H cells transfected with hsa_circ_0000038. (E,F) wound closure assays si were performed using three groups of Huh7 and SK-Hep1 cells (hsa_circ_0000038+miR-92a-2-5p mimics, circ38+miRNC mimics, circNC+miRNC mimics) and three groups of HepG2 and SK-hep1 cells (si-circ38+miR-92a-2-5pinhibitor, si circ38+miRNC inhibitor, siNC+miRNC inhibitor). Co transfection of miR-92a-2-5p mimics andhsa_circ_0000038 partly reverse the effects of single transfection of hsa_circ_0000038. Co-transfection of miR-92a-2-5p inhibitor and si-hsa_circ_0000038 partly reversed the results of single transfection of si-hsa_circ_0000038. (G,H) Transwell invasion assays were performed using the same three groups of these cells. Co-transfection of miR-92a-2-5p mimics and hsa_circ_0000038 could partly reverse the effects of single transfection of hsa_circ_0000038. Co-transfection of miR-92a-2-5p inhibitor and si-hsa_circ_0000038 partly reversed the effects of si-hsa_circ_0000038 transfection. (I,J) The cycle assay showed that the cell cycle shifted from G0/G1 phases to S and G2/M phases after transfecting tumor cells with hsa_circ_0000038 and the direction of the shift begins to reverse after co-transfection with themiR-92a-2-5p inhibitor. Conversely, the cycle was blocked in G0/G1 phases after transfection withhsa_circ_0000038 and the direction of the shift reversed after co-transfection with miR-92a-2-5p mimics.*p<0.05, **p<0.01, ***p<0.001.
Tp53 is a target gene of miR-92a-2-5p: Using the target scan website and Gene Ontology (GO) notation, we identified hsa_circ_0000038 target the downstream p53 pathway (Figure 6A and 6B). The Tp53 mRNA expression levels were significantly lower in 20 HCC patients than in their paired para-neoplastic tissues (Figure 6C) and in four selected hepatocellular carcinoma cell lines compared with HepRG cells (Figure 6D).The same trend was also evident in their protein levels (Figure 6E and 6F). Wild-type Tp53 3'UTR and mutant Tp53 3'UTR were constructed and measured after co transfection with miR-92a-2-5p in 293T cells. Luciferase activity was found to be lost in the wild type (Figure 6G and 6H). In addition, in cells transfected with miR-92a-2-5p mimics, the level of Tp53 mRNA was reduced compared with the NC-transfected group (Figure 6I). Conversely, following transfection with the miR-92a-2-5p inhibitor, Tp53 mRNA exhibited increased expression levels (Figure 6J). This evidence suggests that Tp53 is a direct functional target gene of miR-92a-2-5p in HCC.
Figure 6: Tp53 was a target gene of miR-92a-2-5p.
Note: (A) Potential binding sites of the Tp53gene and miR-92a-2-5p on the website of targets can. The potential binding site information of miR-92a-2-5p’ and Tp53 3-UTR’ is shown in (B) GO notation also suggested that the downstream pathways include the p53 pathway. (C) Tp53 mRNA expression were down regulated in 20 paired HCC tissues and adjacent normal liver tissues. (D) Tp53 mRNA levels were down regulated in liver cancer cell lines compared with HepaRG. (E) Western blot analysis of p53 protein was down regulated in HCC tissue and normal tissue; β-actin was used as a loading control. (F) p53 protein in HCC cells and normal cell in five cell lines. (H) The Tp533-UTR’ wild type site and mutational site. Dual-luciferase reporter assays demonstrated that Tp53 3 UTR’ is a direct target of miR-92a-2-5p. (I) AGO2 confirmed miR-92a-2-5p and Tp53 directly interact. (J,K) The regulation of miR-92a-2-5p can cause the reverse change of Tp53 mRNA expression in cell lines. *p<0.05, **p<0.01,***p<0.001.
Hsa_circ_0000038 Upregulates p53/p21 Expression through miR-92a-2-5p in Liver Cancer Cell Lines
From previous studies, we know that miR-92a-2-5p acts as a cancer switch, but its mechanism in liver cancer has not been studied. We found that miR-92a-2-5p can also promote the proliferation (Figure 7A and 7B) and cell cycle progression (Figure 7C and 7D) of liver cancer cells. Then, we determined whether Tp53 is a downstream target of hsa_circ_0000038 through miR92a-2-5p. First, we conducted rescue assays in HepG2, SK-hep1 and MHCC97-H cells. The CCK-8 assay results showed that the p53 inhibitor pifithrin-αpartially reversed the down-regulation of cell proliferation mediated by the miR-92a-2-5p inhibitor orhsa_circ_0000038 (Figure 7E and 7F). Similar results were obtained in these cell lines in a cell invasion assay (Figure 7G). The RT-qPCR results revealed only minor changes in Tp53 mRNA levels compared with the control group (Figure 7H and 7I). The mismatch phenomenon between the mRNA and protein levels suggests that hsa_circ_0000038 indirectly regulate the expression of Tp53, which confirms our hypothesis that hsa_circ_0000038 may be post-transcriptional level through miR-92a-2-5p to regulatep53/p21 protein expression. The data above suggest that miR-92a-2-5p inhibits the mechanism of Tp53-induced hepato carcinogenesis. Secondly, the results of the western blot analysis showed that the upregulation of hsa_circ_0000038 increased p53/p21 protein expression compared with the control group, while overexpression of miR-92a-2-5p could counteract p53/p21 protein expression (Figure 7J and 7K). In contrast, knockdown of hsa_circ_0000038 could decrease p53/p21 protein expression, but after knock down of hsa_circ_0000038, with continued inhibition of miR-92a-2-5p, the expression of p53/p21proteins were slightly elevated (Figure 7L and 7M). MiR-92a-2-5p levels changed with the expression of hsa_circ_0000038 likewise confirmed this hypothesis. In summary, hsa_circ_0000038 can regulatep53/p21 protein expression through miR-92a-2-5p binding to the 3' - UTR of Tp53 mRNA.
Figure 7: Hsa_circRNA_0000038 promotes the proliferation, migration and invasion of HCC cells by altering miR-92a-2-5p/p53/p21 signals. Note: (A) miR-92a-2-5p mimics could enhance the cell proliferation ability of hepatocellular carcinoma cells. (B) miR-92a-2-5p inhibitor could slow the cell proliferation rate of liver cancer cells. (C,D) miR-92a-2-5p mimics could promote conversion in the cell cycle from the G0/G1 to theS+G2/M phase. After the down regulation of miR-92a-2-5p, this effect reversed. (E–G) pifithrin-α could partially reverse the previous results. (E) Addition of pifithrin-α after transfection of hsa_circ_0000038 partly counter acts the weakening of proliferation by transfected with hsa_circ_0000038 alone. (F) Addition of pifithrin-α after transfection of miR-92a-2-5p inhibitor partly counteract the weakening of proliferation by transfected with miR-92a-2-5p inhibitor alone. (G) Similar remedial effect in invasion assays. (H,I) After transfection of hsa_circ_0000038 (H) or si-hsa_circ_0000038 (I), the Tp53 level was detected by RT-qPCR.(J, K) Protein levels of p53 in HepG2 and MHCC97-H cells were determined by western blot analysis. (J) After co-transfection with hsa_cir_0000038 and miR-92a-2-5p mimics, levels of p53 protein were partly down regulated compared with the levels in the group transfected with si_hsa_cir_0000038 alone? (K) After co-transfection with si-hsa_circ_0000038 and miR-92a-2-5p inhibitor, levels of p53 were partly up-regulated compared with the levels in the group transfected with si-hsa_cir_0000038 alone. (L, M) Protein levels of p53 in HepG2 and MHCC97-H cells were determined by western blot analysis after co transfection with hsa_circ_0000038 and miR-92a-2-5p mimics (L) or co-transfection with si-hsa_cir_0000038 and miR-92a-2-5p inhibitor (M). *p<0.05, **p<0.01, ***p<0.001.
CircRNAs, because of gene trans-editing, possess significant potential for regulating gene expression. CircRNAs are increasingly recognized as novel and unique genetic regulators or biomarkers in the field of cancer research. Dysregulation of their expression or splicing can significantly impact neoplasia development. Several circRNAs are differentially expressed in tumors compared with adjacent non-malignant tissues. They are highly stable and detectable in plasma samples and have been shown to regulate tumor progression by influencing tumor cell cycle, apoptosis, autophagy, immune surveillance effects, vascular regenerative capacity, and cellular energy. In rapidly proliferating tumor cells, circRNAs may not be directly involved in cancer progression, but their expression levels can serve as robust prognostic indicators. The predominant mechanism involves circRNAs effectively modulating the expression and translation of downstream target genes by binding and sequestering miRNAs, thus acting as molecular sponges. One study indicated that hsa_circ_0000038 may be a differential circRNA in colorectal cancer but there are no studies of the function of hsa_circ_0000038.
The database circatlas 3.0 indicated that hsa_circ_0000038 have a lower expression in HCC tissues. RT-qPCR was used to confirm that hsa_circ_0000038 was expressed at a low level in HCC tissues as well as in HCC cell lines. Through functional experiments, we found that over-expression of hsa_circ_0000038 significantly decreased cell viability and proliferation while si-circ_0000038 increased cell viability and proliferation. We confirmed this in vivo through mouse experiments. Together, these results demonstrate that hsa_circ_0000038 serves as a cancer suppressor in HCC progression. MiR-92a-2-5p has been predicted and validated as a target of hsa_circ_0000038. Studies have revealed the tumor accelerator function of miR-92a-2-5p in HCC. To explore the interaction between hsa_circ_0000038 and miR-92a-2-5p, we performed functional experiments and confirmed the hsa_circ_0000038 correlations was between negatively them. correlated First, with miR-92a-2-5p. Second, miR-92a-2-5p mimics significantly increased cell viability and proliferation and promoted the cell cycle, indicating the ability of miR-92a-2-5p to promote HCC cell progression. Lastly, rescue experiments implied that the decrease in cell proliferation due to hsa_circ_0000038 could be recovered by miR-92a-2-5pmimics. These data indicate that hsa_circ_0000038 realized its anti-oncogene function by targeting miR-92a-2-5p. However, the regulation mechanism of this hsa_circ_0000038/miR-92a-2-5p axis is still unknown and needs to be further studied. In HCC, the tumor suppressor p53 inhibits the HCC cell cycle. Importantly, TP53 is predicted to be a putative target gene of miR-92a-2-5p. Therefore, hsa_circ_0000038 influence the regulation of p53 through sponging miR-92a-2-5p thus regulating the cellular functions of HCC. RT-qPCR was employed to test the expression of Tp53 in liver cells transfected with si-circ_0000038 and hsa_circ_0000038 plasmids. The results indicate that overexpression or knockdown of hsa_circ_0000038 did not change the expression levels of Tp53 mRNA. In contrast, western blotting showed that over expression of hsa_circ_0000038 enhanced the p53 protein level, while si hsa_circ_0000038 downregulates the p53 protein level. These data demonstrate that hsa_circ_0000038 plays a vital role inp53 translation but not the transcription. Using rescue assays and western blots, we showed that miR-92a-2-5p could partly reverse the promotion of p53 by hsa_circ_0000038. In general, the present study demonstrates that hsa_circ_0000038 functions as a miR-92a-2-5p sponge and functions by removing the inhibitory effect of miR-92a-2-5p on its target p53, finally, regulating the expression of p53. p21 is considered a translational target of p53 and hsa_circ_0000038 inhibited the p21 protein level through the miR-92a-2-5p/p53 axis (Figure 8).
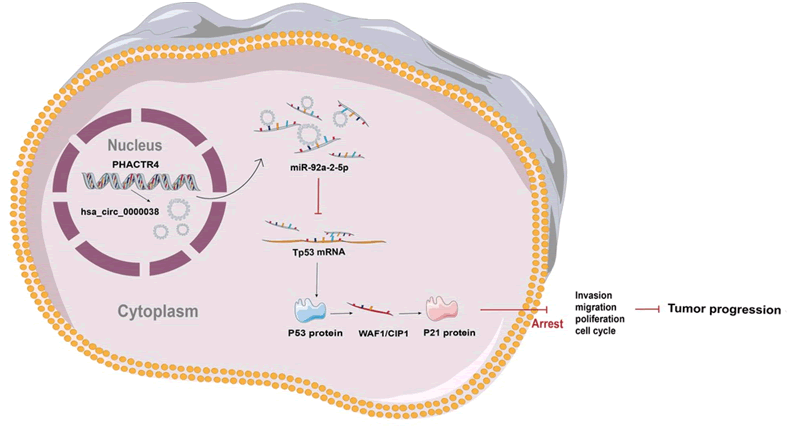
Figure 8: Mechanism of hsa_circ_0000038 function. hsa_circ_0000038 is a novel circRNA derived from the host gene phactr4. hsa_circ_0000038 is transported to the cytoplasm where it sponges miR-92a-2-5p to regulate the p53/p21 axis and thus inhibits cell proliferation, invasion, migration and the cell cycle.hsa_circ_0000038 targets the miR-92a-2-5p/p53/p21 axis to repress the progression of hepato cellular carcinoma.
The TP53 gene is acknowledged as the primary regulator of numerous tumorigenic processes. The relationship between circRNA and P53 has been investigated in various cancers in recent years. For instance; Hsa_circ_0089153 has been demonstrated to target the miR-608/EGFR p53 interaction pathway in ameloblastoma. Circ_0021977 targets the miR-10b-5p/P53/p21 regulatory axis, there by suppressing proliferation, migration and invasion in colorectal cancer. Additionally, Circ_100395 has the ability to inhibit cell growth and metastasis of ovarian cancer cells by regulating the miR-1228/p53/EMT axis. In our study, we presented a new circular RNA (circRNA) hsa_circ_0000038 that targets miR-92a-2-5p/p53 to inhibit liver cancer. The competitive binding mechanism of circular RNA targeting P53 has been found to be a common molecular mechanism for carcinogenesis, as evidenced by our findings in liver cancer. This provides strong evidence that circular RNA is a potential target for tumor treatment. Therefore, hsa_circ_0000038 functions as a p53 accelerator and may be a new diagnostic target and beneficial for HCC patients in the future. The strengths of this study are as follows. First, at both those RNA and protein levels, we validated the association between hsa_circ_0000038 expression and p53. The cellular roles of the p53 gene include regulating the cell cycle, apoptosis, senescence, DNA repair, metabolism and metastasis. Second, a functional enrichment analysis was performed. Third, we show that hsa_circ_0000038 inhibits tumor growth in vivo. These results showed a prominent anti-cancer effect and may guide cancer treatment and targeted drug development. However, there are also limitations. First, this research did not contain independent HCC cohorts. Second, we did not demonstrate the value of significantly elevated hsa_circ_0000038 expression for HCC prognosis. In this research, we finally showed the specific role of hsa_circ_0000038 in HCC and indicated its downstream signaling pathways to determine the biological significance.
The study found that hsa_circ_0000038 is an inhibitory regulator of HCC growth via sponging miR-92a-2-5p to regulate the p53/p21 axis. HCC samples showed low expression of the host gene Phactr4 andhsa_circ_0000038. The hepatoma cell lines Huh7, HepG2 and MHCC-97H had decreased levels ofhsa_circ_0000038. We showed that hsa_circ_0000038 influences p53/p21 levels, hsa_circ_0000038reduces the ability of HCC to proliferate, migrate and invade while accelerating the transition to the G0/G1 phase of the cell cycle. Additionally, miR-92a-2-5p was shown to be the target ofhsa_circ_0000038. These results suggest that hsa_circ_0000038 mediates the miR-92a-2-5p/ p53pathway to regulate HCC progression and proliferation.
J. W. and T. C. Z. designed the study; S. H. Z., and M. T. L., Z. Y. Z. collected the samples and performed the experimental procedures; S. H. Z. and T. C. Z, L.S. performed the data analysis; T. C. Z. and S. H. Z. drafted the manuscript. All authors contributed to and have approved the final manuscript.
This work was supported by grants from National Natural Science Foundation of China (NO.8206080407) and Yunnan Provincial Department of Science and Technology-Kunming Medical University Basic Research Joint Special Project (202101AY070001-283). Young and middle-aged academic and technical leaders reserve talents in Yunnan Province (202205AC160023); Special funds of the Yunnan University “double first-class” construction.
The authors declare no competing financial interests.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Sihang, Luo M, Zeng Z, Shen L, Zou R, et al. (2025) The Novel circRNA hsa_circ_0000038 Inhibits the Progression of Hepatocellular Carcinoma by Sponging miR-92a-2-5p to Regulate the p53/p21 Proteins. Qual Prim Care. 33:51.
Copyright: © 2025 Sihang, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.