Research Article - (2016) Volume 24, Issue 2
Aditya Sood*
Assistant Professor, Chandigarh University, Punjab, India
Ridhi Sood
Assistant Professor, Chandigarh University, Punjab, India
Abhijit Kumar
Assistant Professor, Chandigarh University, Punjab, India
Gaganjot Kaur
Assistant Professor, Chandigarh University, Punjab, India
Candy Sidhu
Assistant Professor, Chandigarh University, Punjab, India
Ved Prakash
Department of biotechnology, College of engineering & Technology, IILM-AHL, Greater Noida, India
Corresponding Author:
Aditya Sood
Department of Preventative Medicines
Northwestern University Feinberg School of Medicine
Chicago, USA
E-mail: er.adityasood@gmail.com
GPCRs are fundamental membrane proteins and they are called as G-Protein Couple Receptors. They are rooted in the cell surface that conveys signals to cell, also known as seven membrane domain receptors. If these receptors do not role correctly, then this may lead to numerous diseases. Orphan Receptors lack endogenous ligands and their particular purpose is not known. Presently there are few ideal structures in databank for following structural biology studies but no orphan GPCR coordinates exist in protein data bank. The aim for gaining appropriate structures for these proteins is to deorphanize them and to study biological applications. There are numerous GPCRs whose 3D structures are obtainable, using those structures as templates, computer based homology modeling method can be used to presume 3D structures of numerous proteins by means of their amino acid sequence. In this study, we have modeled Q99MX9_MOUSE and G2A_ MOUSE, class an orphan GPCRs whose 3D structures have been assumed using Swiss pdb Viewer; the PDB templates used for these orphan GPCRs are 2RH1 and 3UON correspondingly. Based on the outcome of homology modeling of these proteins, their structure has been acquired and their purpose can be presumed on the basis of structural and sequence similarity to known GPCRs. As the structure has been assumed for these GPCRs probable ligands and similarities can be anticipated.
Keywords
GPCRs, Homology Modelling, Structure, Function, Ligand
Introduction
GPCRs are those proteins which are rooted in the surface of the cell which supports in the transmission of the signals every time there is any kind of reaction to nucleotides, proteins amino acids etc.[1-3] Due to their inappropriate functioning, many diseases arise. This makes GPCRs significant drug targets. The superfamily of GPCRs is the major class of surface receptors of cell, they controls diverse tasks of cell for the physiological responses. This makes GPCRs the significant drug objectives and thus is useful in the pharmaceutical expansions. Numerous of the human derived GPCRs are still orphans who are structure and functions or the ligands have yet not identified. [2-5] This makes GPCR significant to the research related to drug designs. Nuclear magnetic resonance spectroscopy and X-ray crystallography are the main techniques used to recognize the structures of proteins. In NMR spectroscopy the proteins are essential not to be crystallized but dissolved protein with high applications are required. Therefore, GPCR structures cannot be acknowledged with this method. X-ray crystallography is responsible for the atomic information of the proteins which are globular. But GPCRs structures cannot be resolved from this technique because it is very hard to crystallize them.[6,7] Therefore, different structure based methods have to be tracked to resolve the structures and functions of GPCRs. Building models by aligning the target sequences and studying their evolutionary relationships can assistance in formative the structures and functions of GPCRs.[8-10]
Software’s and Data sources
• GLIDA: GPCR-Ligand Database
• pBLAST
• Protein data Bank (PDB)
• SWISS-PROT
• Swiss pdb Viewer
• SAVS: Structural Analysis and Verification Server
• Computed Atlas of Surface Topography of proteins (CASTp)
Need of Project
In situation of the human genome, GPCRs are the main gene superfamily. Maximum of the drug based research is going on in this field because not working of these receptors lead to numerous diseases. Considerate their structures, functions and ligand binding sites can aid in drug design which can aid in dealing with the diseases. Deorphanization of orphan GPCRs can widen research and help in discovering new drug targets and also aid in understanding the functioning of the orphan GPCRs on cellular level, as they are involved in numerous signaling pathways.
Methodology
1. Two orphan GPCRs – Q99MX9_mouse and G2A_mouse were selected using GLIDA.
2. The fasta files of these two orphan G-proteins were retrieved.
3. Protein Blast was done and subsequent hits were obtained.
4. The pdb ID’s of different templates of interest that are 2RH1 and 3UON were fetched and downloaded using PDB.
5. The fasta sequences of the orphan G-proteins and their respective templates were uploaded in swiss-prot.
6. Homology Modelling was done and the resulting modeled proteins were saved as .pdb files.
7. The energies were checked using swiss pdb viewer. For the structure to be in stable state its energy should be minimum.
8. The ramachandran plots were studied for the modeled proteins and the residues residing in favorable and allowed regions were studied using Structural Analysis and Verification Server.
9. The residues residing in disallowed regions were shifted to allowed regions via loop modeling and
10. Subsequently energies were computed and the modeled structures were checked for the same.
11. The active sites or ligand binding sites were predicted using CASTp server.
12. The fasta sequences of the modeled orphan G proteins were saved and used as an input format in BLAST.
13. Phylogenetic analysis was done.
14. On the basis of Phylogenetic analysis, the possible functions were predicted.
Results and Discussions
All the biological data and the chemical information of the ligands of GPCRs are obtainable in the GPCR-Ligand Database well-known as GLIDA. It includes both bioinformatics and chemo informatics approaches to examine the data of GPCRs [Figure 1-4]. Consequently, it is beneficial in the drug related finding. Mostly the GPCR data is connected to human, mouse and rats. Mouse and rats is the perfect organisms used for the discoveries of numerous drugs. [9-11]

Figure 1: Result for Query Q99MX9_MOUSE.
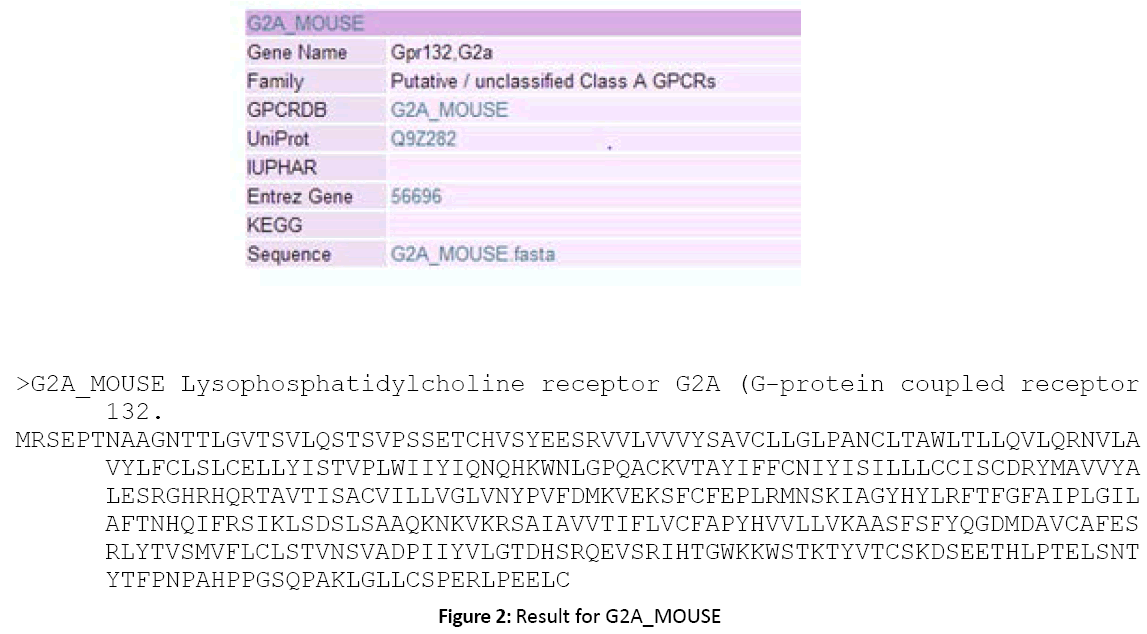
Figure 2: Result for G2A_MOUSE
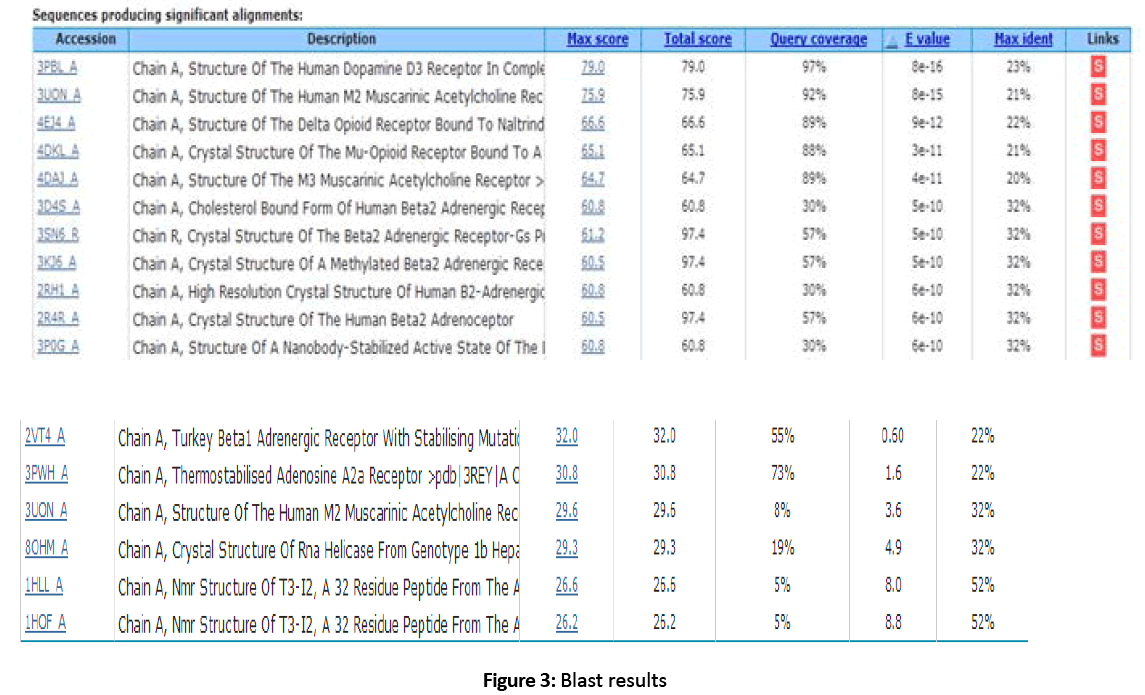
Figure 3: Blast results
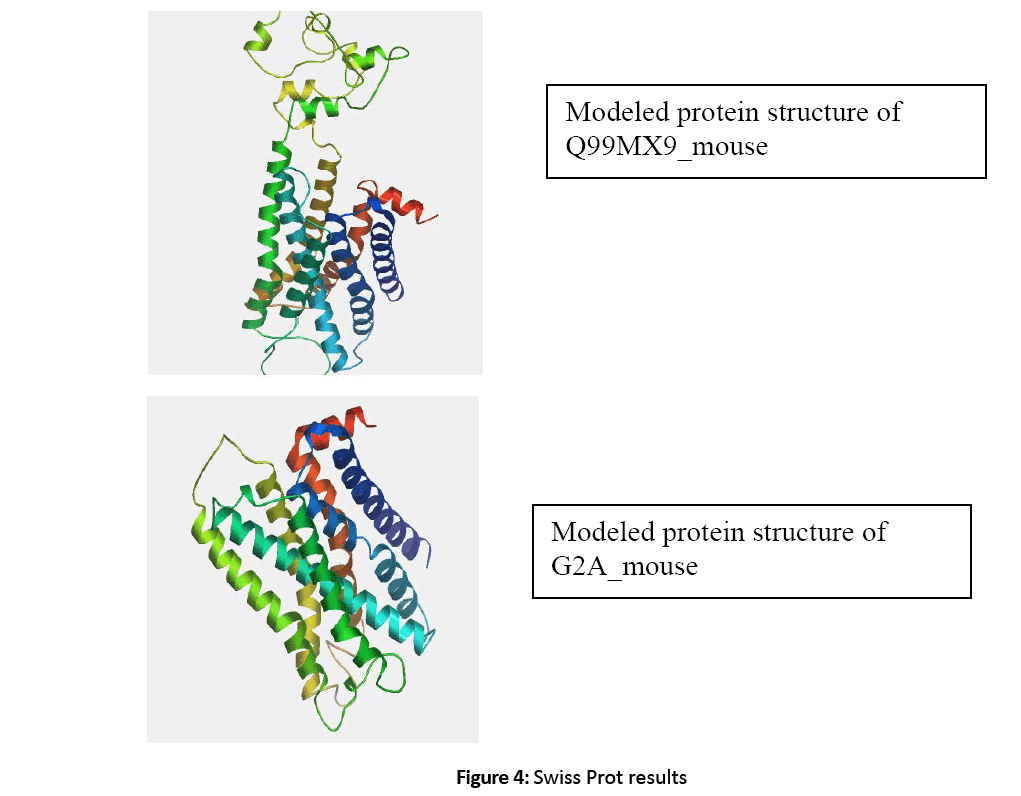
Figure 4: Swiss Prot results
All the data related to ligands such as the active sites, the binding pockets, the charge on the ligands, the volume of the binding pockets (aids in determining the size of ligands) can assistance in calculating the related drugs [Figure 5-7]. Mouse Models are extensively used in drug discovery and clinical trials. If structural and functional information of mouse orphan GPCR can be gained, it can be used in deducing active sites or ligand binding sites for numerous drugs [Figure 8-11]. This is a stepping stone in finding drugs for numerous diseases in humans as mouse and human genetic makeup is very similar. Employing the information about a deorphanized GPCR also will offer useful insights into the purposes related to the protein and various pathways complex in the cells.[10-12]
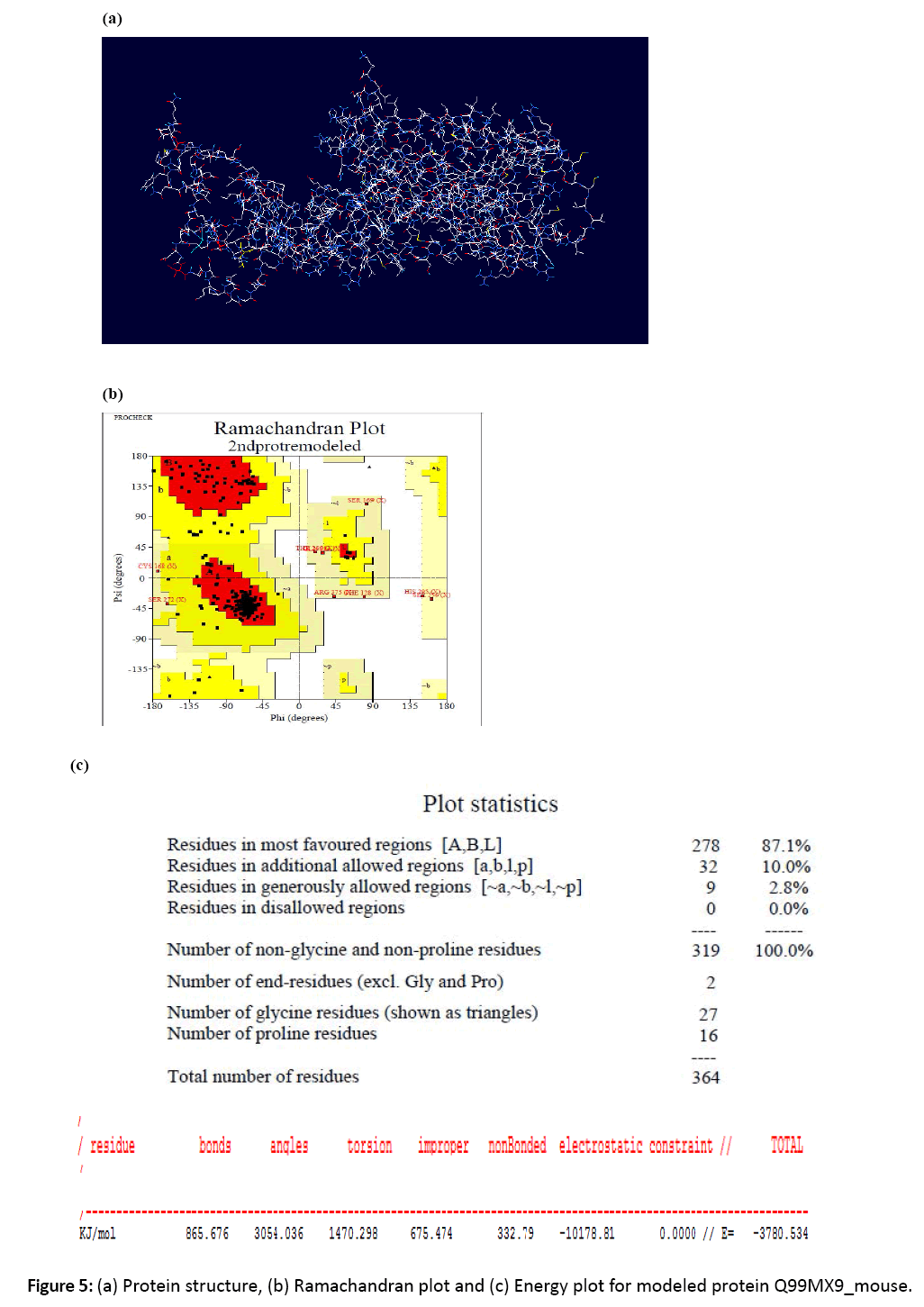
Figure 5: (a) Protein structure, (b) Ramachandran plot and (c) Energy plot for modeled protein Q99MX9_mouse.
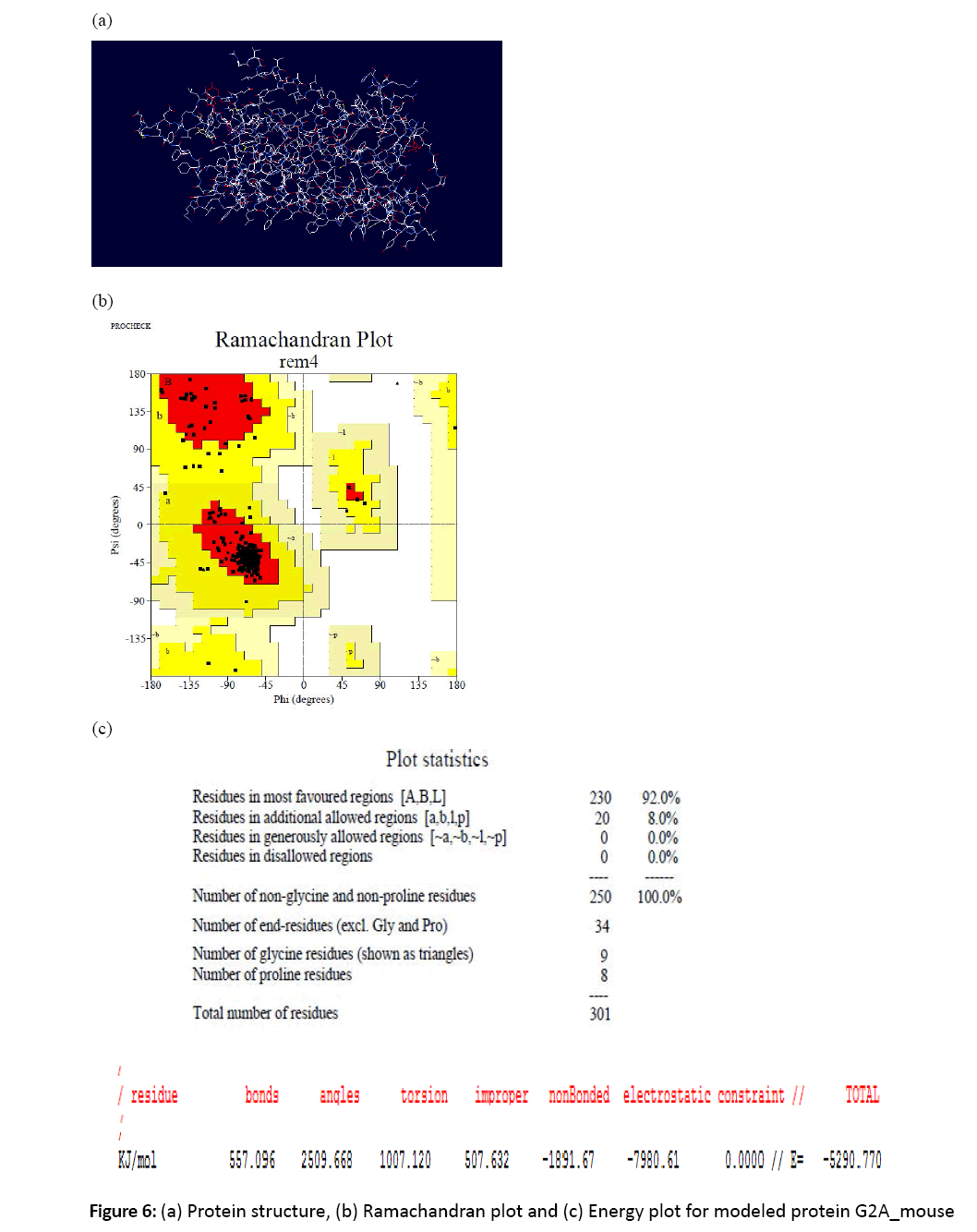
Figure 6: (a) Protein structure, (b) Ramachandran plot and (c) Energy plot for modeled protein G2A_mouse

Figure 7: Fasta file of modeled protein Q99MX9_mouse.

Figure 8: Fasta file of modeled protein G2A_mouse.
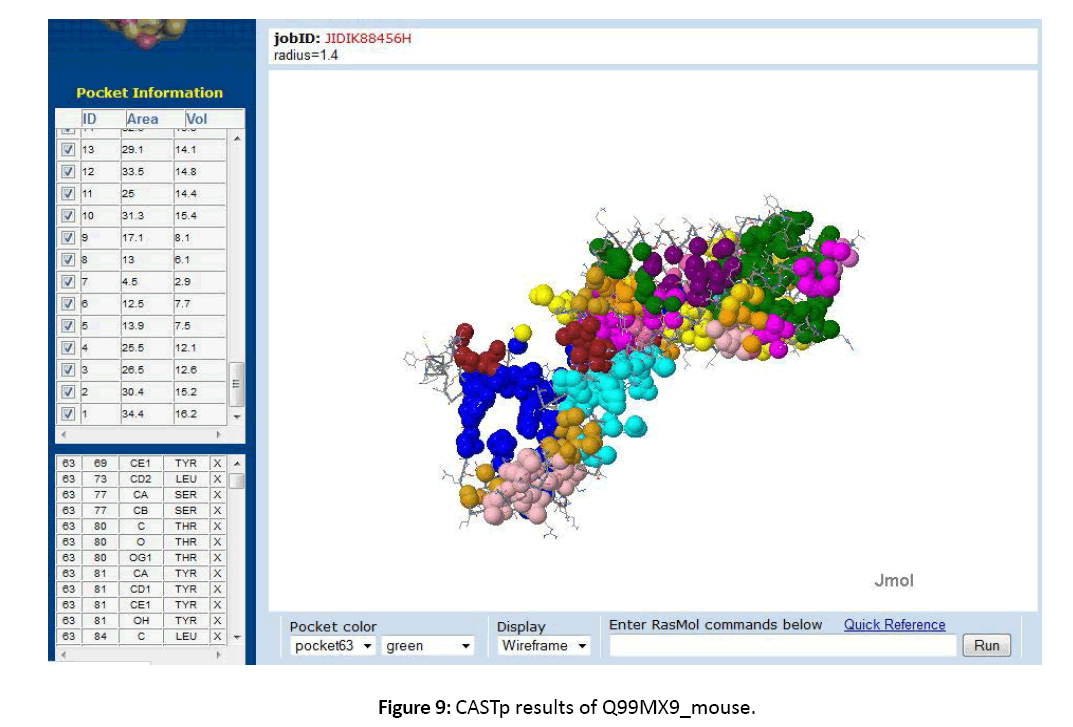
Figure 9: CASTp results of Q99MX9_mouse.
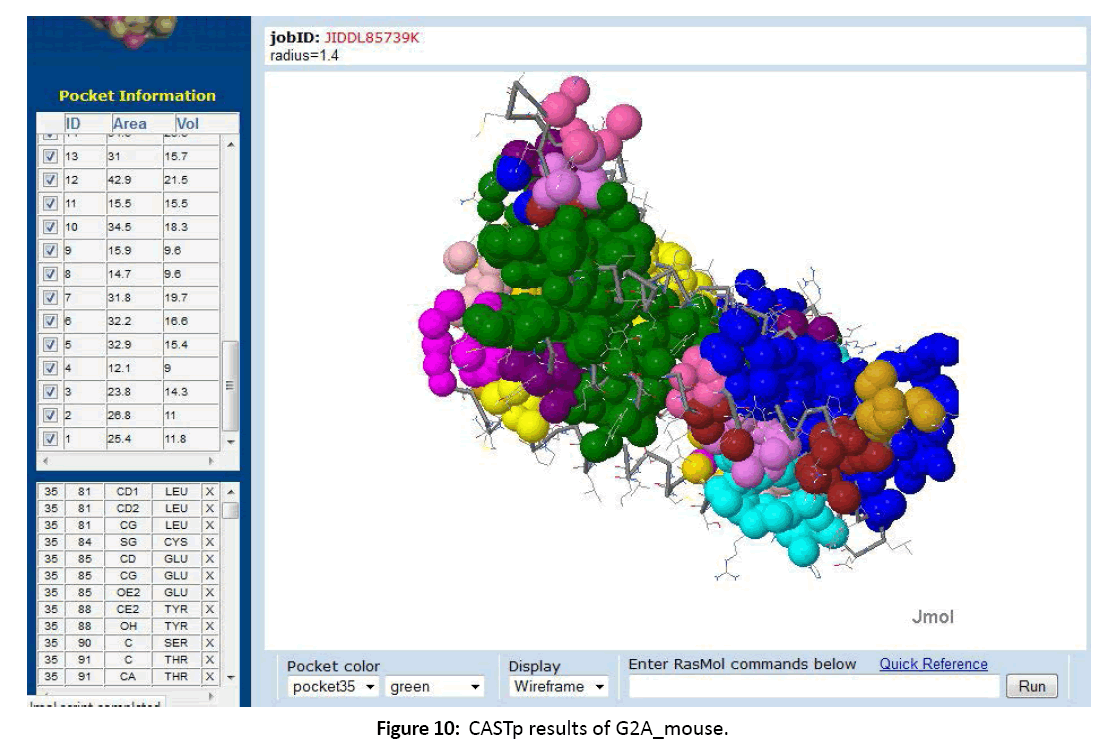
Figure 10: CASTp results of G2A_mouse.
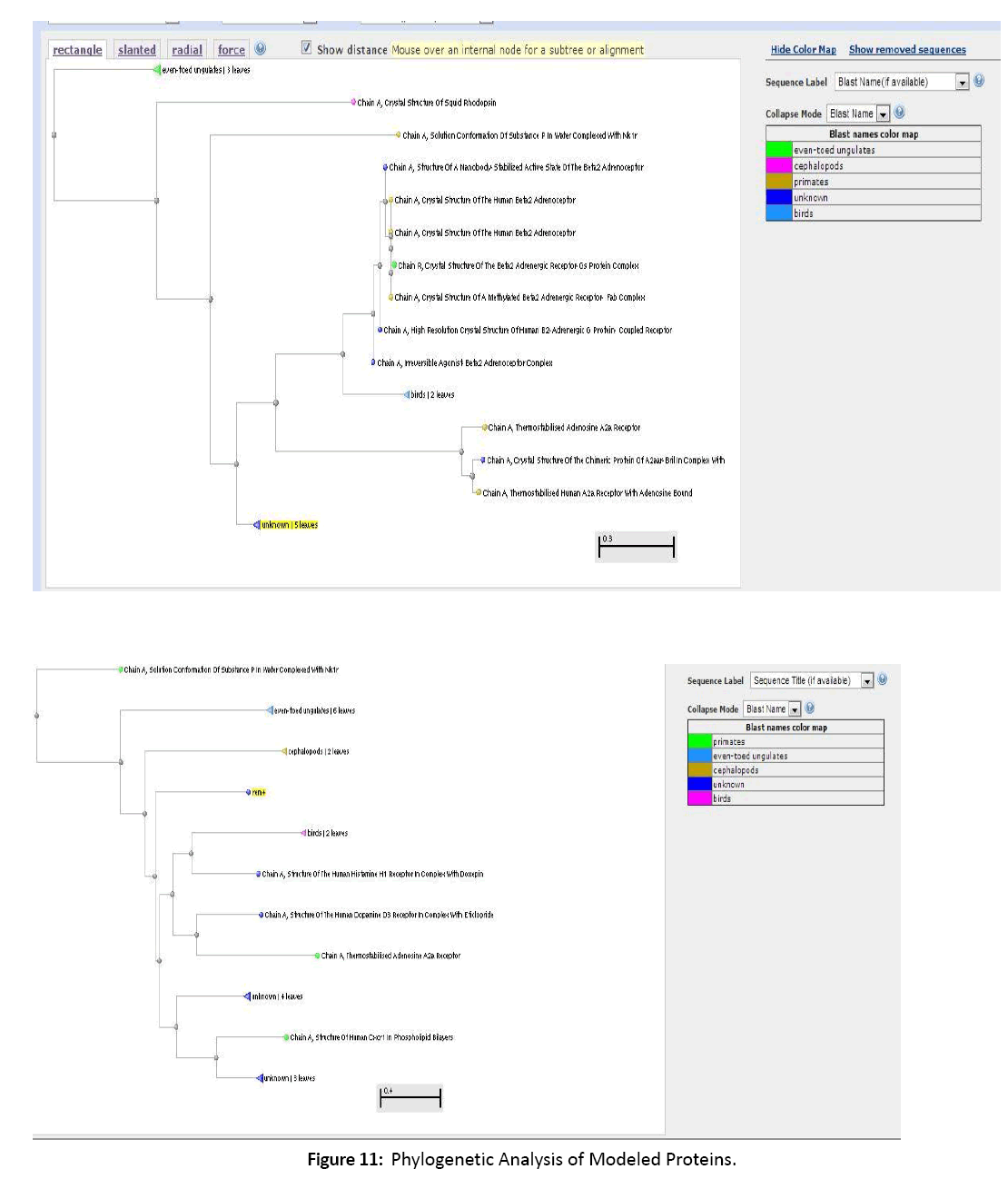
Figure 11: Phylogenetic Analysis of Modeled Proteins.
The Phylogenetic examination aided in understanding and predicting the purposes of modeled proteins. The closely connected proteins display a important use in drug discovery, ligand prediction and additional pharmacological use. Hence we conclude that the modeled G-proteins play a vital role in pharmacological science and predicting specific ligands.[10-12]