Keywords
Endothelial growth factor; Polymorphism; Congenital heart defect
Introduction
Congenital heart defect (CHD) is one of the most common birth deformities, with approximately 0.6-0.8% of live infant births receiving the diagnosis [1]. Despite this prevalence, the pathogenesis of CHD remains unknown. However, a small number of CHD cases are caused by a single gene mutation or other chromosomal aberrations, leaving the remaining 90% CHD diagnoses resulting from a heterogenous etiology including a variety of genetic factors and environmental factors [2].
The heart is the first organ to form and function during development [3]. Numerous signaling pathways contribute to its development, and include vascular endothelial growth factor (VEGF), GATA4, and Nkx2.5 [4-6]. Of these pathways, VEGF signaling has been shown to be linked to CHD, with its spatiotemporal expression pattern indicating a potential role [7]. The VEGF gene is located on chromosome 6p12 and consists of eight exons, which can be alternatively spliced to form a family of proteins [8].
It has been reported that VEGF is required for proper heart morphogenesis at stages [7]. Additionally, VEGF-expressing endothelial cells located in the cushion-forming region may be a unique subpopulation of endothelial cells that are predetermined to transform from endocardium to mesenchyme (EMT) [9]. Importantly, maintaining an appropriate timing and dosage of VEGF during heart development has been shown in animal models to be shared in various cardiovascular developmental defects. This work includes work with transgenic mice heterozygous for the VEGF allele, which showed a two- to three-fold increase in VEGF levels [10]. Past work has also shown that increased VEGF levels during the development of the right ventricular outflow tract can lead to abnormal development of both cushion and myocardial structures [11].
VEGF gene polymorphisms may play a role in susceptibility to congenital valvuloseptal heart defects. It has also been reported that VEGF genetic polymorphisms may also be associated with CHD, including tetralogy of fallot (TOF) [12] and ventricular septal defect (VSD) [13].
Materials and Methods
Identification and eligibility of relevant studies
We carried out an online search in PubMed and Web of Science databases for related articles published before March 31, 2016 using the following terms: “congenital heart defects or congenital heart diseases or heart, malformation of heart abnormalities or CHD” and “mutation or polymorphism or variation” and “vascular endothelial growth factor or VEGF”. To expand the range of our studies, we also used the same terms in Chinese to search the Chinese National Knowledge Infrastructure (CNKI), Wangfang Database, and Chinese Biology medicine disc (CBM). References of the retrieved articles were also scanned for additional studies.
We included case-controls with human subjects that studied the relationship between VEGF C2578A, G1154A, and G634C mutations and CHD susceptibility in both English and Chinese languages. All phenotypes of CHD, including ventricular septal defect, patent formen ovale, atrial septal defect, patent ductus arteriosus, and coarctation of the aorta were included in this meta-analysis. However, CHD patients who had additional congenital, co-morbid anomalies such as Down syndrome were excluded [17]. Research articles utilizing animal subjects, reviews, commentaries, case reports, and unpublished reports were also excluded [18-20]. Studies that did not provide raw data of allele frequencies in the initial publications were excluded [12], though we attempted to obtain primary data by writing to the authors. Finally, when the research populations overlapped, we avoided repetition by including only the research with the broadest data set for the meta-analysis.
Data extraction
All data were collected independently by two authors (Zhang and Mo) and any discrepancy was resolved by a third co-author (Yu). The following information was collected or counted from each study: first author, year of publication, country of origin, ethnicity, type of CHD, number of cases and controls, counts of alleles in case and control groups in case-control studies, and Hardy-Weinberg equilibrium (Tables 1-3).
Statistical analysis
STATA (version 11.0; StataCorp, College Station, Texas, USA) was used for meta-analysis. All gene models for the VEGF C2578A, G1154A, and G634C mutations were estimated. The existence of heterogeneity between studies was ascertained using a Q-statistic. The pooled odds ratio (OR) was evaluated with models based on either fixed or random effects assumptions. A random effects model was used if a significant Q statistic (P<0.1) indicated heterogeneity in the studies. In all other cases a fixed effects model was used. The 95% confidence interval (CI) of OR was also computed. The distributions of genetypes in the controls were checked for Hardy-Weinberg equilibrium. Begg’s and Egger’s tests were used to assess the publication bias and P<0.05 was considered as statistically significant. Funnel plots of the VEGF C2578A (for C versus A ), G1154A (for G versus A), and G634C (for G versus C) were performed to look for evidence of publication bias. The funnel plot should be asymmetric in the case of publication bias and symmetric in the case of no publication bias.
Results
Characteristics of eligible studies
Figure 1 shows the literature retrieval and research selection processes. We found that 6 articles containing 1080 cases and 2289 controls were relevant to C2578A, 4 articles containing 528 cases and 1036 controls were relevant to G1154A, and 6 articles containing 1081 cases and 2281 controls were relevant to G634C. A total of 4 articles explored a single type of CHD [13,14,21,22], whereas various types of CHD were included in the other two articles [2,23]. Among them, three studies were carried out in China [13,21,24], one in Chile [14], one in Germany [22], and one in the Netherlands [24]. The distributions of the genotypes in the control groups were consistent with Hardy-Weinberg equilibrium (p>0.05) in all six studies in which the control groups were representative (Tables 1-3).
| Reference |
Country of origin |
Ethnicity |
Type of CHDs |
Sample size
(Case/Control) |
Cases |
Controls |
HWE |
| CC |
CA |
AA |
CC |
CA |
AA |
| Calderon (2009) |
Chile |
American |
All types |
61/61 |
14 |
32 |
15 |
16 |
33 |
12 |
0.50 |
| Smedts et al. [24] |
Netherland |
European |
All types |
187/307 |
53 |
88 |
44 |
71 |
153 |
88 |
0.77 |
| Xie et al. [13] |
China |
Asian |
VSD |
222/352 |
124 |
83 |
15 |
211 |
124 |
17 |
0.82 |
| Stalmans et al. [22] |
Germany |
European |
All types |
58/316 |
12 |
28 |
18 |
97 |
157 |
62 |
0.91 |
| Wang et al. [21] |
China |
Asian |
All types |
238/134 |
135 |
85 |
18 |
66 |
59 |
9 |
0.38 |
| Gu [23] |
China |
Asian |
All types |
316/557 |
272 |
182 |
22 |
305 |
225 |
27 |
0.073 |
HWE: Hardy-Weinberg equilibrium; VSD: ventricular septal defect
Table 1: Characteristics of the included studies on VEGF C2578A polymorphism and congenital heart defects (CHD).
| Reference |
Country of origin |
Ethnicity |
Type of CHDs |
Sample size
(case/control) |
Cases |
Controls |
HWE |
| GG |
GA |
AA |
GG |
GA |
AA |
| Calderon (2009) |
Chile |
American |
All types |
61/61 |
29 |
26 |
6 |
37 |
19 |
5 |
0.27 |
| Smedts et al. [24] |
Netherland |
European |
All types |
185/312 |
90 |
79 |
18 |
134 |
130 |
43 |
0.21 |
| Xie et al. [13] |
China |
Asian |
VSD |
222/352 |
156 |
60 |
6 |
255 |
89 |
8 |
0.94 |
| Stalmans et al. [22] |
Germany |
European |
All types |
58/316 |
16 |
32 |
10 |
152 |
132 |
32 |
0.67 |
HWE: Hardy-Weinberg equilibrium; VSD: Ventricular septal defect
Table 2: Characteristics of the included studies on VEGF G1154A polymorphism and congenital heart defects (CHD).
| Reference |
Country of origin |
Ethnicity |
Type of CHDs |
Sample size
(case/control) |
Cases |
Controls |
HWE |
| CC |
CA |
AA |
CC |
CA |
AA |
| Calderon (2009) |
Chile |
American |
All types |
61/61 |
27 |
28 |
26 |
27 |
26 |
8 |
0.66 |
| Smedts et al. [24] |
Netherland |
European |
All types |
184/303 |
77 |
85 |
22 |
131 |
133 |
39 |
0.57 |
| Xie et al. [13] |
China |
Asian |
VSD |
222/352 |
78 |
118 |
26 |
68 |
181 |
103 |
0.47 |
| Stalmans et al. [22] |
Germany |
European |
All types |
58/316 |
30 |
25 |
3 |
147 |
135 |
34 |
0.72 |
| Wang et al. [21] |
China |
Asian |
All types |
240/135 |
90 |
116 |
34 |
50 |
63 |
22 |
0.77 |
| Gu [23] |
China |
Asian |
All types |
476/557 |
153 |
248 |
75 |
183 |
283 |
91 |
0.29 |
HWE: Hardy-Weinberg equilibrium; VSD: ventricular septal defect
Table 3: Characteristics of the included studies on VEGF G634C polymorphism and congenital heart defects (CHD).
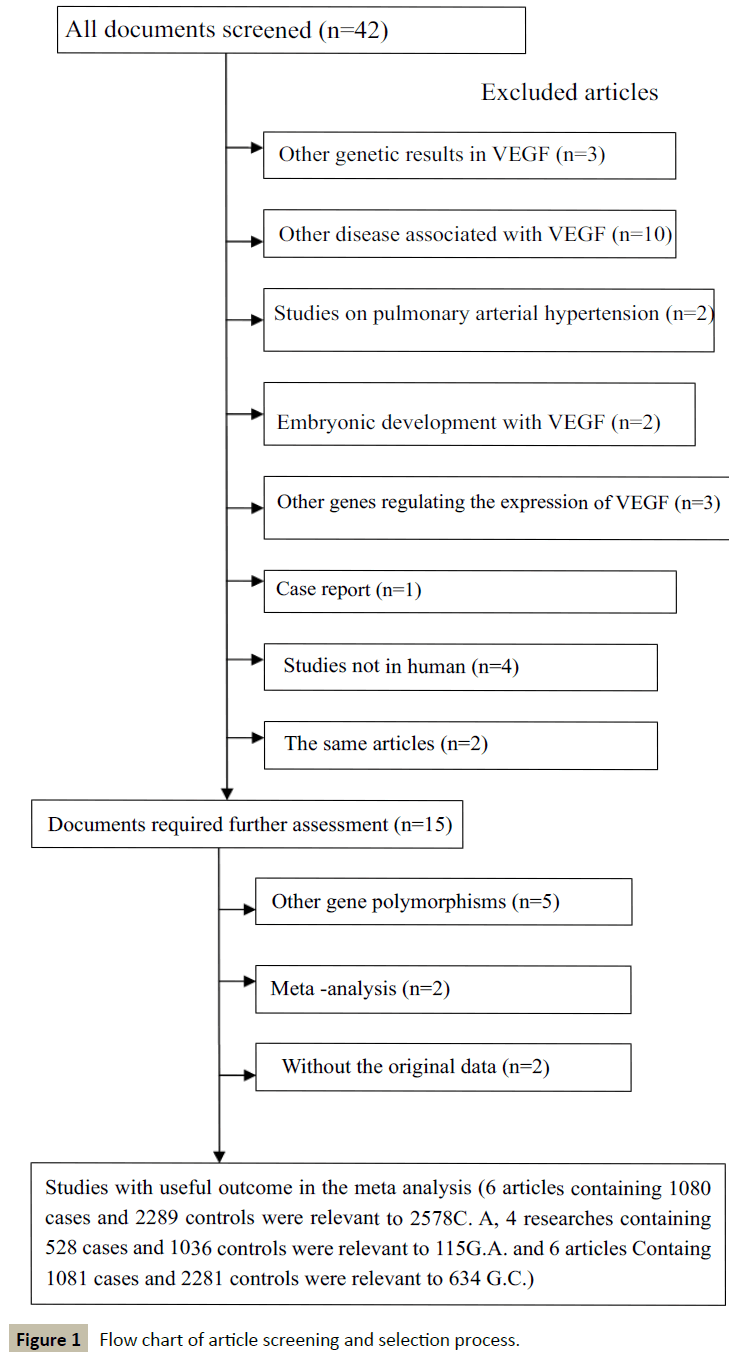
Figure 1: Flow chart of article screening and selection process.
Results of the meta-analysis
We accessed the full genotype distributions and observed that the VEGF C2578A polymorphism was unrelated to CHD in allelic comparisons (A vs. C: OR=1.016, 95% CI: 0.851, 1.214; Pheterogeneity=0.072), homozygote comparison (AA vs. CC: OR=1.048, 95% CI: 0.792, 1.386; Pheterogeneity=0.134, Figure 2), dominant model (AA/AC vs. CC: OR=0.950, 95% CI: 0.812, 1.112; Pheterogeneity=0.177), and recessive model (AA vs. AC/CC: OR=1.080, 95% CI: 0.842, 1.385; Pheterogeneity=0.314). Moreover, no significant associations were found in G1154A allelic comparison (A vs. G: OR=1.202, 95% CI: 0.837, 1.725; Pheterogeneity=0.011), homozygote comparison (AA vs. GG: OR=1.302, 95% CI: 0.596, 2.844; Pheterogeneity=0.036, Figure 3), dominant model (AA/AG vs. GG: OR=1.310, 95% CI: 0.843, 2.036; Pheterogeneity=0.020), and recessive model (AA vs. AG/GG OR=1.078, 95% CI: 0.630, 1.846; Pheterogeneity=0.196). We also found no relationship in G634C alleleic comparison with susceptibility to CHD (C vs. G: OR = 0.850, 95% CI: 0.659, 1.097; Pheterogeneity=0.000), homozygote comparison (CC vs. GG: OR=0. 633, 95% CI: 0.357, 1.123; Pheterogeneity=0. 000, Figure 4), dominant model (CC/CG vs. GG: OR=0.849, 95% CI: 0.629, 1.145; Pheterogeneity=0.009), and recessive model (CC vs. CG/GG: OR=0.675, 95% CI: 0.435, 1.046; Pheterogeneity=0.007).
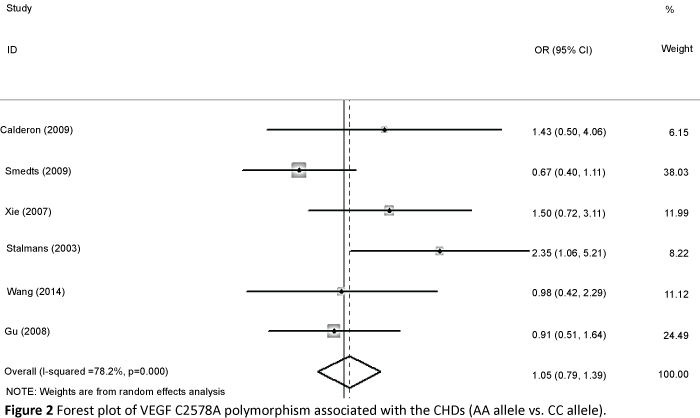
Figure 2: Forest plot of VEGF C2578A polymorphism associated with the CHDs (AA allele vs. CC allele)
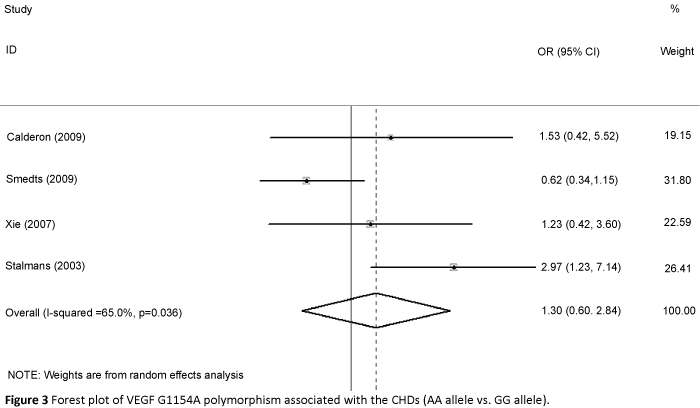
Figure 3: Forest plot of VEGF G1154A polymorphism associated with the CHDs (AA allele vs. GG allele).
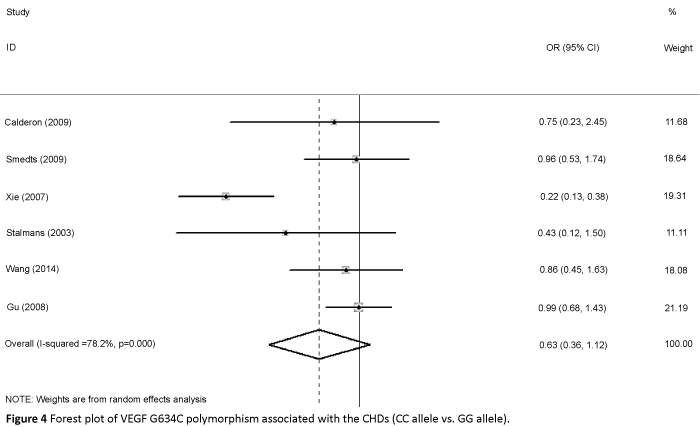
Figure 4: Forest plot of VEGF G634C polymorphism associated with the CHDs (CC allele vs. GG allele).
Galbraith plot
We then created a Galbraith plot to graphically assess the sources of heterogeneity of VEGF G634C (Figure 5). Only one study was identified as the main source of heterogeneity [8]. After the outlier study was excluded, we still did not find a connection in G634C allelic comparison and CHD (C vs. G: OR=0.979, 95% CI: 0.866, 1.108; Pheterogeneity=0.974 ), homozygote comparison (CC vs. GG: OR=0. 908, 95% CI: 0.694, 1.188; Pheterogeneity=0. 787 Figure 6), dominant model (CC/CG vs. GG: OR=1.004, 95% CI: 0.842, 1.197; Pheterogeneity=0.953 ), and recessive model (CC vs. CG/GG: OR=0.890, 95% CI: 0.696, 1.137; Pheterogeneity=0.817 ).
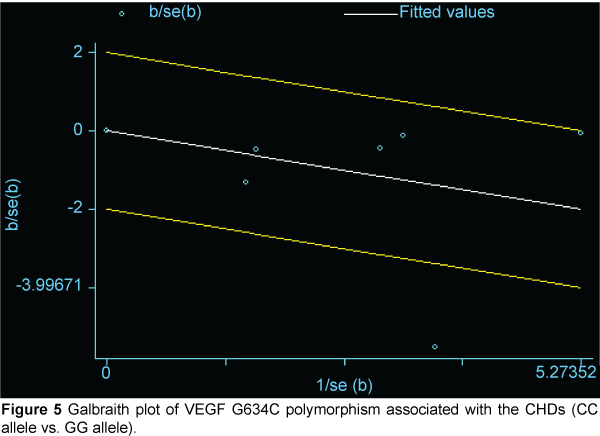
Figure 5: Galbraith plot of VEGF G634C polymorphism associated with the CHDs (CC allele vs. GG allele).
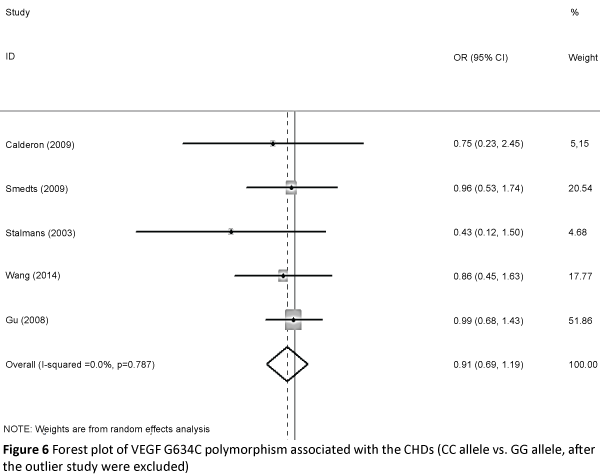
Figure 6: Forest plot of VEGF G634C polymorphism associated with the CHDs (CC allele vs. GG allele, after the outlier study were excluded).
Publication bias
Publication bias was assessed using both a Begg's funnel plot and Egger's test. Begg's funnel plots are shown in Figure 7 (P=0.536 for VEGF C2578A C allele versus A allele), Figure 8 (P=1.000 for VEGF G1154A G allele versus A allele), and Figure 9 (P=1.000 for VEGF G634C G allele versus C allele). Egger's test was then performed for to determine any publication bias (P=0.891 for A allele versus C allele, P=0.687 for A allele versus G allele, P=0.910 for C allele versus G allele). No publication biases were found.
Figure 7: Funnel plot in the meta-analysis of the association between VEGF C2578A polymorphism and the risk of CHDs.
Figure 8: Funnel plot in the meta-analysis of the association between VEGF G1154A polymorphism and the risk of CHDs.
Figure 9: Funnel plot in the meta-analysis of the association between VEGF G634C polymorphism and the risk of CHDs.
Discussion
VEGF is located on chromosome 6q21. 33 and includes eight exons. Alternative splicing can result in several different protein isoforms [8]. Past work has shown that some of VEGF polymorphisms may be associated with differential VEGF expression in vitro [25]. Among these, polymorphisms in the VEGF promoter region (0.2578 C>A, rs 69994) and located on VEGFI exon 634 (G>C, rs 2010963) may be associated with the 3’ noncoding region [26]. Other studies have also implicated these VEGF mutations disease states, including in CHD. For instance, Vannay et al. [6] found that VEGF polymorphism 634C (+ 405 c) increased the risk of CHD, while Lambrecht et al. [12] showed that VEGF haplotype 2578A/1154A/634G significantly reduced the risk of tetralogy of fallot (TOF), a form of congenital heart disease.
Griffin et al. [15] was the first to perform a study on the relationship between VEGF C2578A, G1154A and G634C polymorphisms and the risk of CHD. The results of this work indicated that there was no relationship between these three polymorphisms and CHD. Li et al. [16] found that the allele, genotype, and haplotype of VEGF were identified with an association for susceptibility to CHD. Furthermore, that there were differences between CHD with or without DiGeorge syndrome; namely, that specific haplotypes (CGC) had significant protective effects for reducing the risk for CHD in a non-DiGeorge syndrome population. Given these discrepancies in the literature, the purpose of this meta-analysis was to assess whether or not there was an association between VEGF C2578A, G1154A, and G634C polymorphisms and the risk for CHD.
In this meta-analysis, our results showed that none of the VEGF C2578A, G1154A, and G634C polymorphisms were significantly associated with risk for CHD. However the studies of VEGF C2578A and G1154A polymorphisms were too few to sufficient exam heterogeneity. Therefore, we adopted a Galbraith plot to assess the sources of heterogeneity of VEGF G634C, and eliminated one study based on the result. We conducted our statistical analysis again, but still failed to find a relationship between VEGF G634C polymorphism and CHD. One reason for this failure could be that only six studies were included in this analysis and that our statistical power was too low to allow for robust statistical conclusions. Moreover, there is significant difference in the genetic background, exposure to environmental factors, and risk factors in life styles between Asian, American, and European populations. Since our meta-analysis included all three of these populations, it is possible that this diversity masked any significant findings.
Future work will need to address some of the limitations present in this meta-analysis. First, all of the studies were carried out using only four different countries, including only Asian, American, and European populations. Second, most of the studies selected grouped all heart defects together. Uniform deÂnitions and categories of CHDs might be needed in later investigations to parse out more specific genetic contributions to each type of CHD. Third, recent work has shown that peri-conceptional use of multivitamins containing folic acid can reduce the incidence of CHD [19,27]. As we did not assess the folate intake of the populations in question, it will be important to include this variable in future work as a potential factor in the prevention of CHDs. Finally, the influence of other environmental factors, such intrauterine infection as well as high doses of radioactive material and/or drugs should also be taken into consideration. Despite these limitations, our meta-analysis offers more evidence for the association (or lack thereof) between VEGF C2578A, G1154A, and G634C gene polymorphisms and the risk of CHD. Collectively, future studies using larger samples and better-matched controls will be needed to further confirm the findings from our metaanalysis.
Conclusion
This meta-analysis did not provide evidence for an association between VEGF C2578A, G1154A, and G634C genetic polymorphisms and CHD risk. These results do not support the hypothesis that VEGF C2578A, G1154A, and G634C polymorphisms may be a susceptibility marker of CHD. However, larger-sized sample studies will be needed in the future to validate our findings. Additionally, other factors such as plasma homocysteine levels, enzymatic activity, parental genotypes, and vitamin complex intake will also need to be included. Finally, more gene-gene and gene-environment interaction studies will be needed in future work, which should lead to a better and more comprehensive understanding of the association between VEGF polymorphisms and CHD risk.
Role of Funding Source
This work was supported by funding from the National Natural Science Foundation of China (No. 81370279).
References
- Hoffman JI, Kaplan S (2002) The incidence of congenital heart disease. J Am Coll Cardiol 39: 1890-1900.
- Basinko A, Giovannucci Uzielli ML, Scarselli G, Priolo M, Timpani G, et al. (2012) Clinical and molecular cytogenetic studies in ring chromosome 5: report of a child with congenital abnormalities. Eur J Med Genet 55: 112-116.
- Paige SL, Plonowska K, Xu A, Wu SM (2015) Molecular regulation of cardiomyocyte differentiation. Circ Res 116: 341-353.
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, et al. (2003) GATA4 mutations cause human ongenital heart defects and reveal an interaction with TBX5. Nature 424: 443-447.
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, et al. (1998) Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281: 108-111.
- Vannay A, Vásárhelyi B, Környei M, Treszl A, Kozma G, Györffy B (2006) Single-nucleotide polymorphisms of VEGF gene are associated with risk of congenital valvuloseptal heart defects. Am Heart J 151: 878-881.
- Dor Y, Camenisch TD, Itin A, Fishman GI, McDonald JA, et al. (2001) A novel role for VEGF in endocardial cushion formation and its potential contribution to congenital heart defects.Development 128: 1531-1538.
- Vincenti V, Cassano C, Rocchi M, Persico G (1996) Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3.Circulation 93: 1493-1495.
- Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A (1999) Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Dev Biol 212: 307-322.
- Lambrechts D, Carmeliet P (2004) Genetics in zebrafish, mice, and humans to dissect congenital heart disease: insights in the role of VEGF. Curr Top Dev Biol 62: 189-224.
- Van den Akker NM, Molin DG, Peters PP, Maas S, Wisse LJ, et al. (2007) Tetralogy of fallot and alterations in vascular endothelial growth factor-A signaling and notch signaling in mouseembryos solely expressing the VEGF120 isoform. Circ Res 100: 842-849.
- Lambrechts D, Devriendt K, Driscoll DA, Goldmuntz E, Gewillig M, et al. (2005) Low expression VEGF haplotype increases the risk for tetralogy of Fallot: a family based association study. J Med Genet 42: 519-522.
- Xie J, Yi L, Xu ZF, Mo XM, Hu YL, et al. (2007) VEGF C-634G polymorphism is associated with protection from isolated ventricular septal defect: case-controland TDT studies. Eur J Hum Genet 15: 1246-1251.
- Calderón JF, Puga AR, Guzmán ML, Astete CP, Arriaza M, et al. (2009) VEGFA polymorphisms and cardiovascular anomalies in 22q11 microdeletion syndrome:a case-control and family-based study. Biol Res 42: 461-468.
- Griffin HR, Hall DH, Topf A, Eden J, Stuart AG, et al. (2009) Genetic variation in VEGF does not contribute significantly to the risk of congenital cardiovascular malformation. PLoS One 4: e4978.
- Li Yi fei, Zhou Kaiyu, Xie Liang, Wang Chuan, Tang Haowen, et al. (2012) A meta-analysis of the association between vascular endothelial growth factor gene polymorphisms and congenital heart disease. Chin J Evid Based Pediatr 10: 3969.
- Ackerman C, Locke AE, Feingold E, Reshey B, Espana K, et al. (2012) An excess of deleterious variants in VEGF-A pathway genes in Down-syndrome-associated atrioventricular septaldefects. Am J Hum Genet 91: 646-659.
- Basinko A, Giovannucci Uzielli ML, Scarselli G, Priolo M, Timpani G, et al. (2012) Clinical and molecular cytogenetic studies in ring chromosome 5: report of a child with congenital abnormalities. Eur J Med Genet 55: 112-126.
- Frigerio A, Stevenson DA, Grimmer JF (2012) The genetics of vascular anomalies. Curr Opin Otolaryngol Head Neck Surg 20: 527-532.
- Zhou J, Pashmforoush M, Sucov HM (2012) Endothelial neuropilin disruption in mice causes DiGeorge syndrome-like malformations via mechanisms distinctto those caused by loss of Tbx1. PLoS One 7: e32429.
- Wang E, Wang Z, Liu S, Gu H, Gong D, et al. (2014) Polymorphisms of VEGF, TGFβ1, TGFβR2 and conotruncal heart defects in a Chinese population. Mol Biol Rep 41: 1763-1770.
- Stalmans I, Lambrechts D, De Smet F, Jansen S, Wang J, et al. (2003) VEGF: a modifier of the del22q11 (DiGeorge) syndrome? Nat Med 9: 173-182.
- Gu H (2008) Gene polymorphisms and risk of congenital heart disease in a Chinese Population: [D]. Nanjing: Nanjing medical university.
- Smedts HP, Isaacs A, de Costa D, Uitterlinden AG, van Duijn CM, et al. (2010) VEGF polymorphisms are associated with endoc ardial cushion defects: a family-based case-control study. Pediatr Res 67: 23-28.
- Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, et al. (1999) Novel polymorphisms in the promoter and 5' UTR regions of the human vascular endothelial growth factor gene. Hum Immunol 60: 1245-1249.
- Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE (2000) Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation withvariation in VEGF protein production. Cytokine 12: 1232-1235.
- Van den Dungen JJ, Karliczek GF, Brenken U, van der Heide JN, Wildevuur CR (1983) The effect of prostaglandin E1 in patients undergoing clinical cardiopulmonary bypass. Ann Thorac Surg 35: 406-414.