Research Article - (2017) Volume 3, Issue 3
Jean-Claude Perez*
Retired Interdisciplinary Researcher (IBM), PhD Maths § Computer Science, France
*Corresponding Author:
Jean-Claude Perez
Retired Interdisciplinary Researcher (IBM)
PhD Maths § Computer Science
7 Avenue de Terre-rouge F33127 Martignas Bordeaux Metropole, France.
Tel: 33 0781181112
E-mail: jeanclaudeperez2@gmail.com, jeanclaudeperez3@free.fr
Received date: July 17, 2017; Accepted date: July 31, 2017; Published date: August 04, 2017
Citation: Perez JC (2017) The “Master Code of DNA”: Towards the Discovery of the SNPs Function (Single-Nucleotide Polymorphism). J Clin Epigenet. 3:26. doi: 10.21767/2472-1158.100060
Background: Every human individual is differentiated from all other humans by the few million SNPs. We are interested in the immediate neighborhood of each SNP. Would the SNP point have particular properties with respect to the surrounding nucleotides at short or medium distance?
Methods: The regions encompassing each SNP are analyzed by the Biomathematical method of the "Master Code of DNA" published elsewhere. In particular, we will use a progressive approach of dichotomy type.
Results: There are then two types of remarkable results. Evidence of fractal properties such as self-similarity and scale invariance. On the other hand, the position of each SNP seems to play a functional role of the "active site" type as it is found in genes and proteins.
Conclusions: Each SNP would be more important by its precise location in the genome than by the value of its local mutation T/C or A/G for example. Consequently, SNPs would play a major functional role.
Keywords
Human genome; Fractals; Self similarity; Scale invariance
Introduction
“Fourth, we need to turn our new genomic information into an engine of pharmaceutical discovery. Individuals human differ from one another by about one base pair per thousand. These “Single Nucleotide Polymorphisms” (SNPs) are markers that can allow epidemiologists to uncover the genetic basis of many diseases. Moreover, the analysis of SNPs will provide us with the power to uncover the genetic basis of our individual capabilities such as mathematic ability, memory, physical coordination, and even, perhaps, creativity.” By Professor David Baltimore (Nobel prizewinner) in the historical Human Genome issue of NATURE [1].
Since 1990, we are looking for possible hidden numerical codes that would control and structure the DNA of genes and genomes [2-4]. On the one hand, all the genetic difference between two human beings comes from some 1% of TCAG bases of the whole human genome constituting the SNPs [5,6]. On the other hand, we have discovered and published a law and a method of analysis: the "master code of DNA" [7,8], which UNIFIES all the information of the 3 types of CODES of Biology: DNA, RNA and amino acid sequences. Other numerical codes of DNA and genomes were discovered during these 25 years [8-11]. Some of these codes, as will be the case here, are fractal [12-16].
Methods
Master code summary
Starting from the atomic masses constituting nucleotides and amino acids, a numerical scale of integers characterizing each bio atom, each TCAG DNA base, each UCAG RNA base, or each amino acid, an integer numbers scale code is obtained. Then, for each sequence of double - stranded DNA to be analyzed, the sequence of integers that characterizes it (genomics) is constructed as well as the sequence of amino acids that would encode this double strand if each of the strands was a potential protein (proteomics). The remarkable fact is that this proteomics image still exists, even for regions not translated into proteins (junk DNA). The computational methodology of the Master code [7,8] then produces 2 patterned images (2D curves, Figure 1) which are very strongly correlated. This would mean that beyond the visible sequence of DNA there would be a kind of Master Code being manifested by two supports of biological information: the sequences of DNA and of amino acids, the RNA image constituting a kind of neutral element like the zero of the mathematics. Our thousands of genes and genomes Master Code analyses (viruses, archaeas, bacteria, eucharyotes) demonstrated [7,8] that the extremums (max and min) signify functional regions like proteins active sites, fragility points like chromosomes breakpoints). The main discovery of the paper will be the fact that each SNP is located precisely in such extremums. This allows us to consider the probable functionality of each SNP.
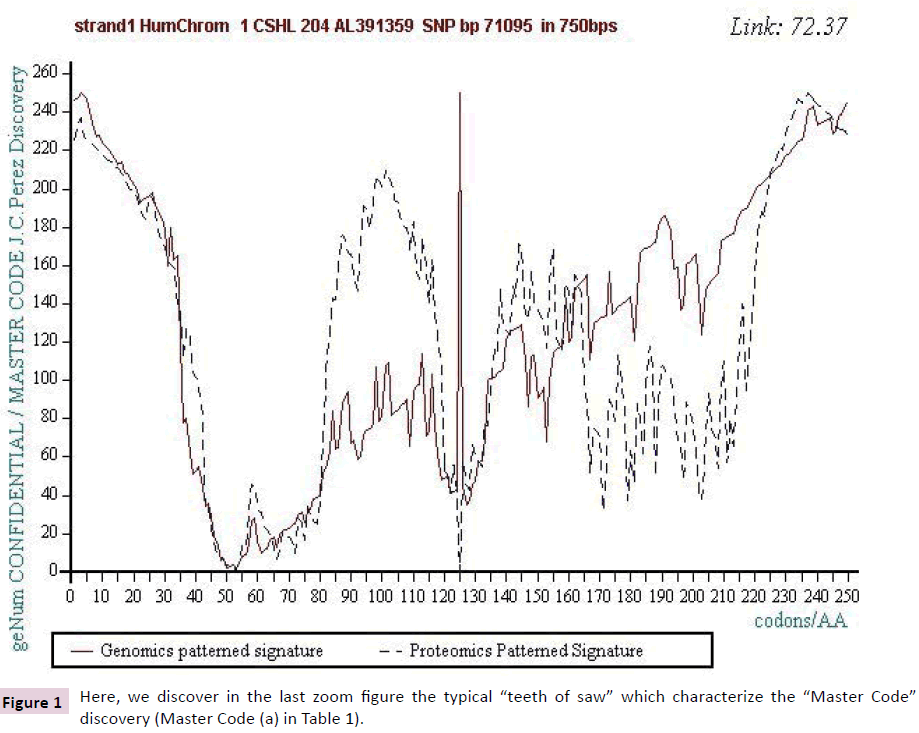
Figure 1: Here, we discover in the last zoom figure the typical “teeth of saw” which characterize the “Master Code” discovery (Master Code (a) in Table 1).
Figure 1 below illustrates such a Master Code computing in the case of the SNP referenced (a) in Table 1. On the one hand, there is a strong correlation between the 2 images Genomics and Proteomics (72.37%) and on the other hand the kinds of "saw teeth" characteristic of fractal roughness’s [12,13]. This "saw teeth" discovery is at the origin of the resonance and period’s research to be published [13].
| SNP | dbSNP | Chrom | Band | GenBank ver | Golden path contig |
SNP pos relative to | ||
|---|---|---|---|---|---|---|---|---|
| GB | Contig | Chrom (kb) | ||||||
| …/... | ||||||||
| TSC1004969 | 2827238 | Chr1 | - | AL391359.6 | ctg12483 | 71095 | 47949 | 75163.3(a) |
| …/... | ||||||||
| TSC1004975 | 2827244 | Chr1 | - | AL391359.6 | ctg12483 | 151041 | 93765 | 75209.2(c) |
| …/... | ||||||||
| TSC1270753 | - | Chr1 | - | AL391359.5 | ctg12483 | 58750 | 119856 | 75235.3(b) |
| …/… | ||||||||
Table 1: Chromosome 1 (“Single Nucleotide Polymorphisms” (SNPs)).
We work with the public SNPs web database from “CSHL” https://www.snp.cshl.org/. We proceed by successive analyzes of dichotomous type (embedded zooms): a sequence of 12000 bases is defined on each side of the SNP, then a second halfreduced sequence (6000 bases) is analyzed, then 3000 bases, then 1500 bases, and finally 750 bases (Figures 2-11).
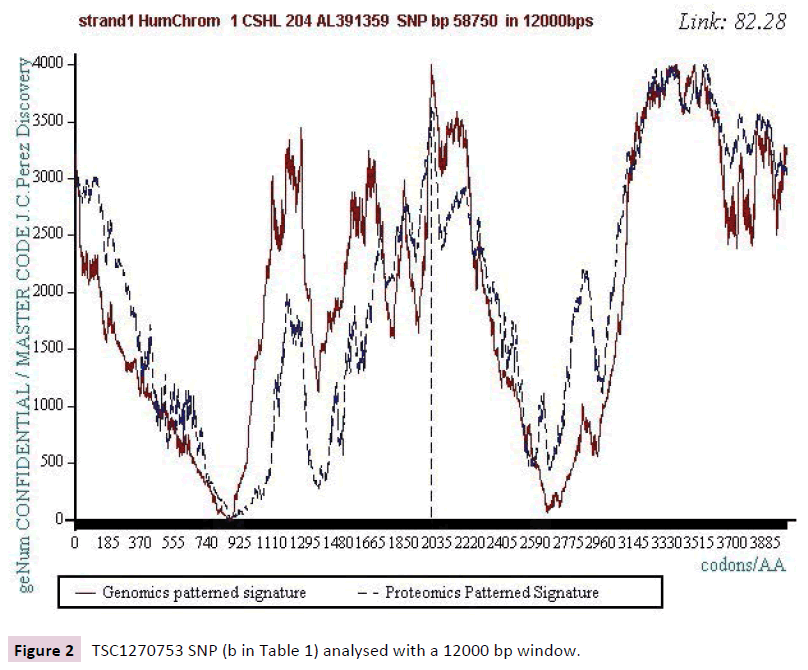
Figure 2: TSC1270753 SNP (b in Table 1) analysed with a 12000 bp window.
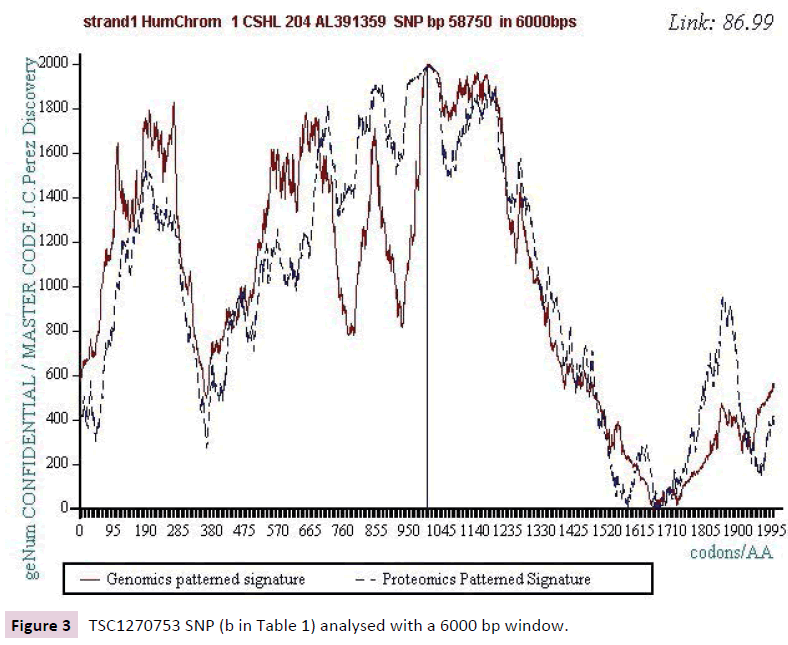
Figure 3: TSC1270753 SNP (b in Table 1) analysed with a 6000 bp window.
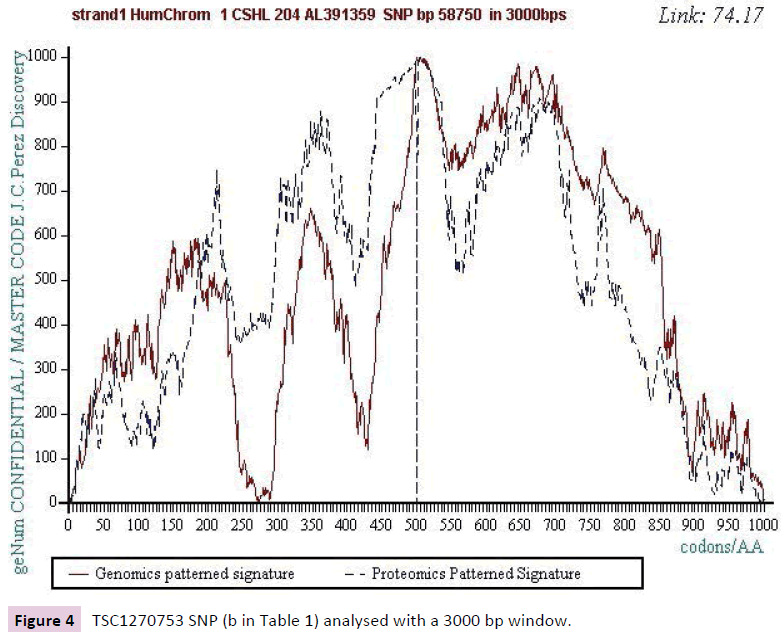
Figure 4: TSC1270753 SNP (b in Table 1) analysed with a 3000 bp window.
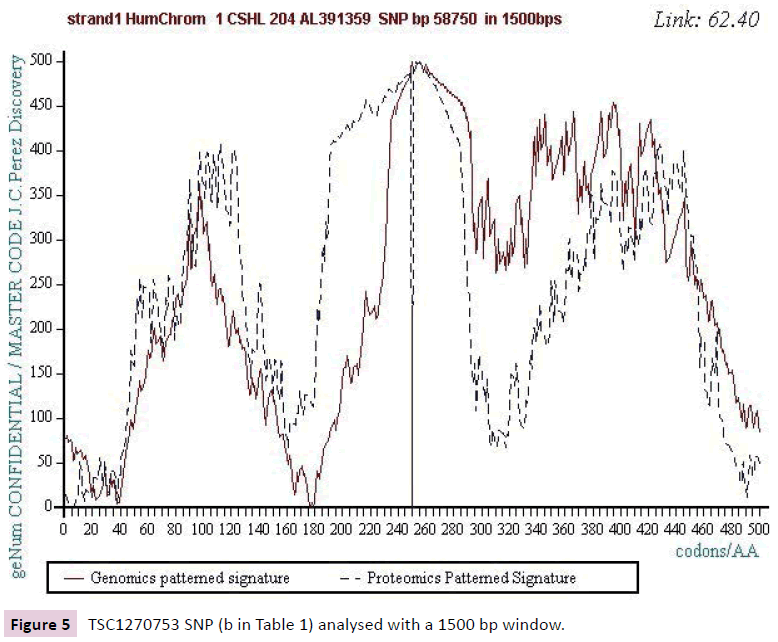
Figure 5: TSC1270753 SNP (b in Table 1) analysed with a 1500 bp window.
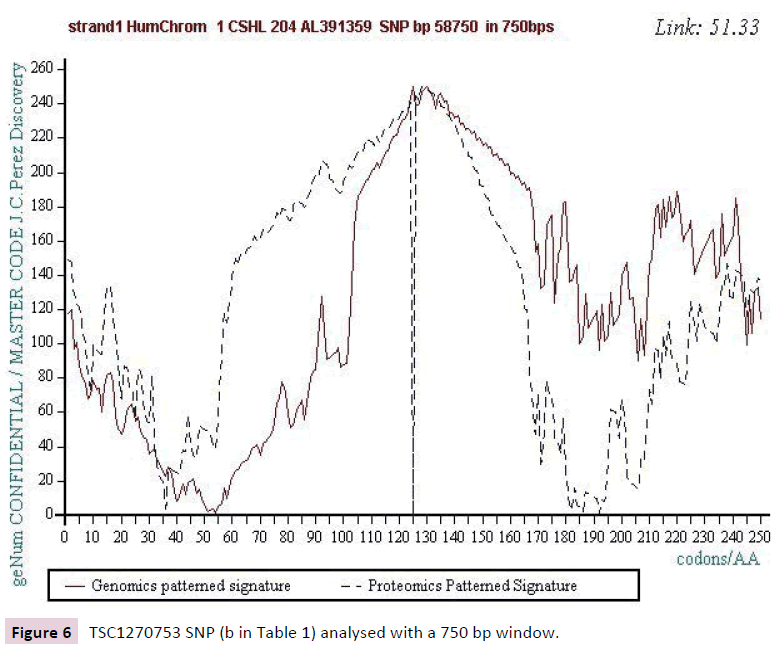
Figure 6: TSC1270753 SNP (b in Table 1) analysed with a 750 bp window.
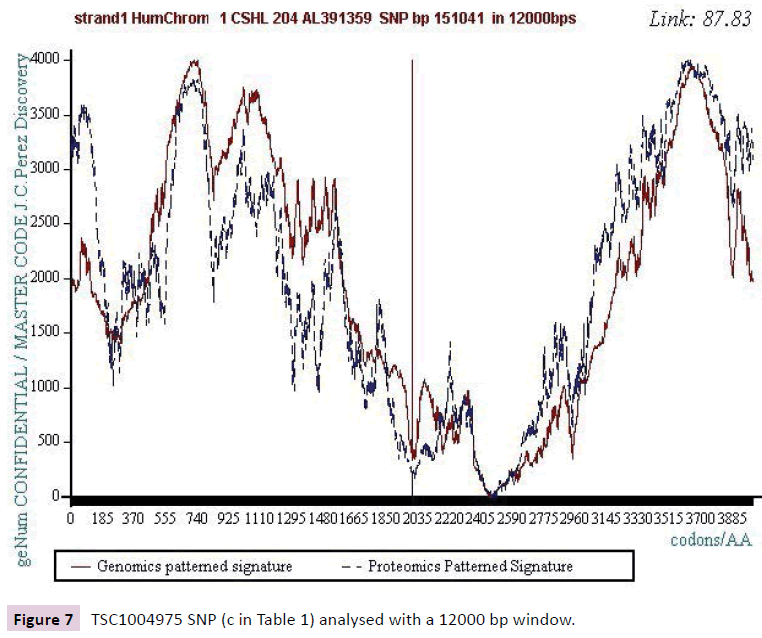
Figure 7: TSC1004975 SNP (c in Table 1) analysed with a 12000 bp window.
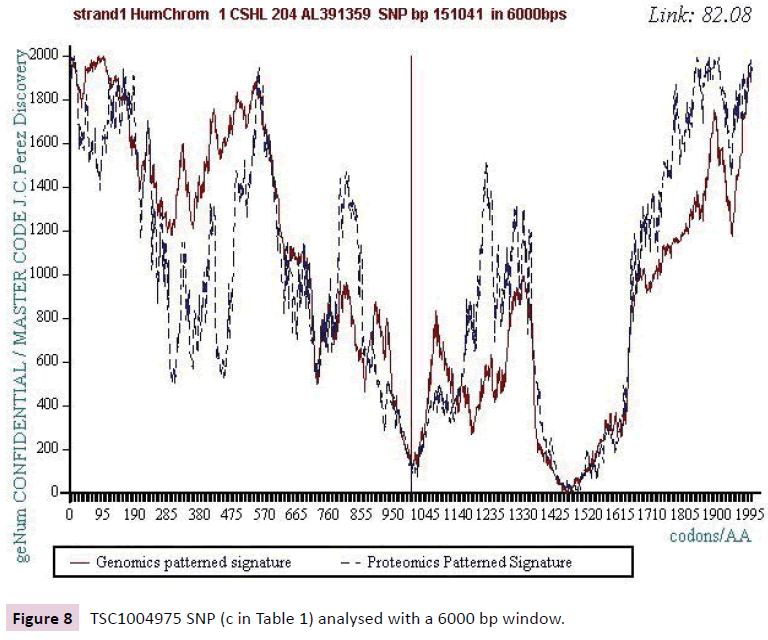
Figure 8: TSC1004975 SNP (c in Table 1) analysed with a 6000 bp window.
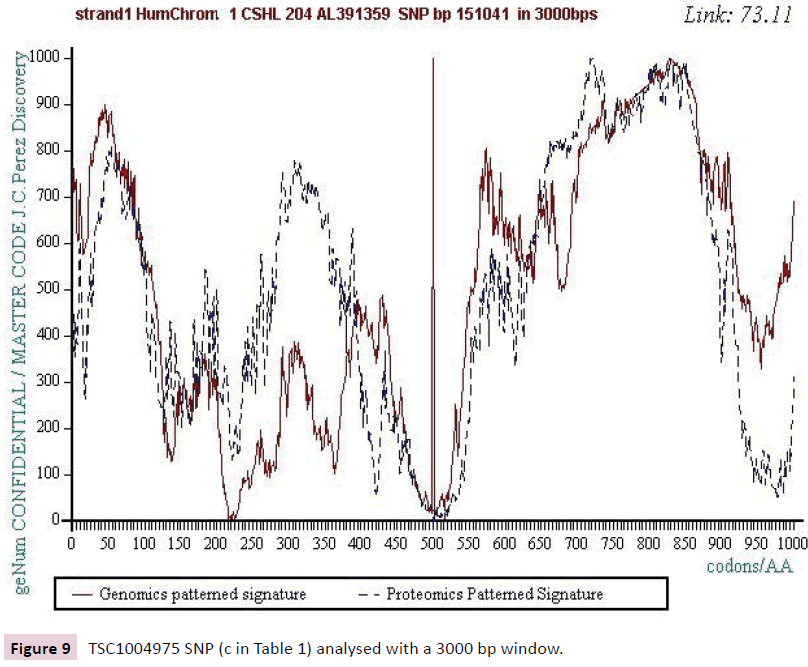
Figure 9: TSC1004975 SNP (c in Table 1) analysed with a 3000 bp window.
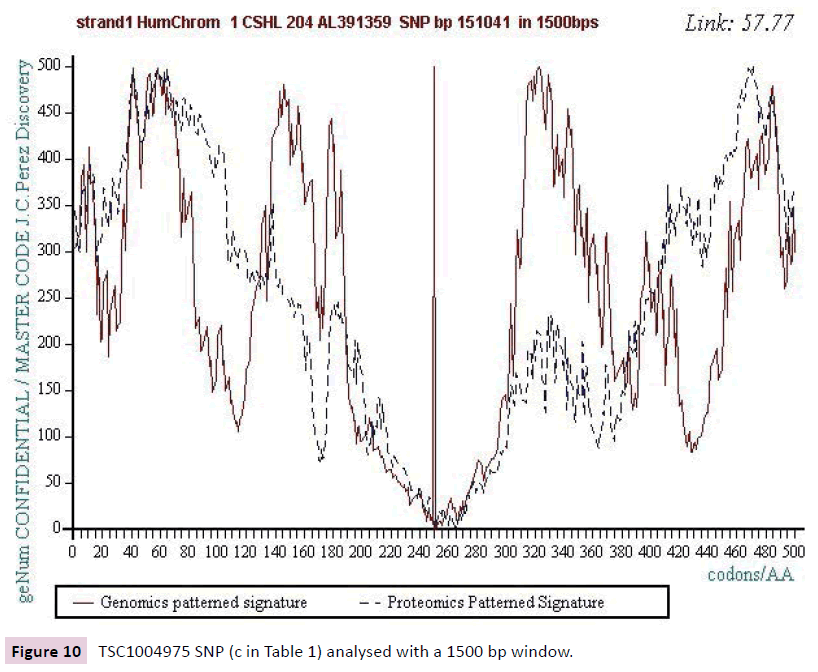
Figure 10: TSC1004975 SNP (c in Table 1) analysed with a 1500 bp window.
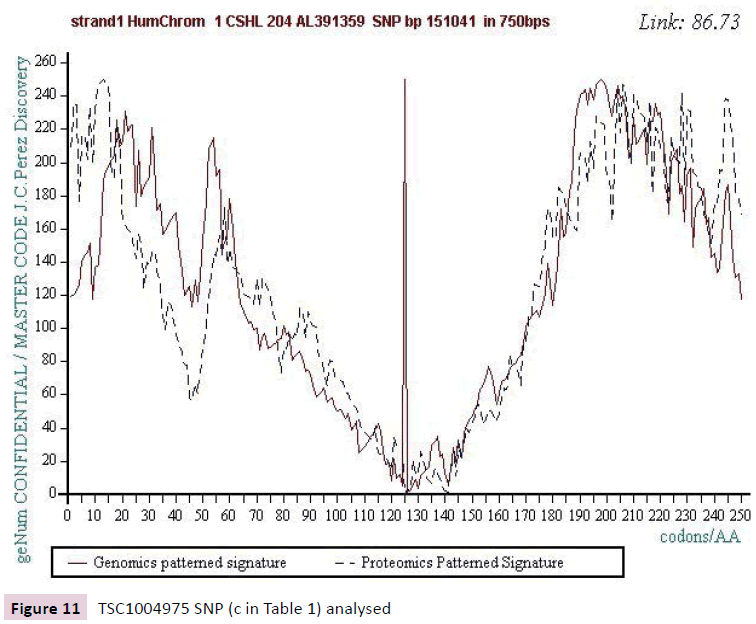
Figure 11: TSC1004975 SNP (c in Table 1) analysed.
Results
We analyzed several tens of SNPs according to this Master Code of DNA method. We have included only the two most representative types in this synthesis document, knowing however that ALL the cases analyzed lead to results of this nature. Analyzing a first SNP by the Analyzing a first SNP by the "Master code" (b, figures 2-6), then Analysing a second SNP by the “Master Code”... (c, figures 7-11). So, generalizing this type of analyses reveals the evidence of this astonishing law: “in the human genome, it would seem that SNPs are more important by their position than by their value. It is, probably, this position located in strategic functional sites which “amplifies “the scope of effect and the global differentiation of these simple local specific changes which are SNPs single mutations".
Discussion
Collection and the availability of thousands of SNPs of the human genome (https://SNP.cshl.org et https://www.ncbi.nlm.nih.gov) OPEN very great QUESTIONS like:
• Do SNPs have an influence at long distance in the genome (stability etc.)?
• Do SNPs play crucial part promoters, modulation, tuning in the function and expression of genes?
• Do the SNPs play on the contrary a local role and at very short distance? etc.
We studied this problem according to various points of view. With the Master Code:
• Local short distance analysis (900 to 1000 bases available around each SNP from data bases).
• Analysis of very large contigs multi-SNP: example of the study of 120,000 bases containing 16 SNPs which one changes overall swapping them on their extreme nucleotides values).
• Research of the scope of effects of the SNP on the characteristic GENOMIC CODE of the studied areas (this paper)
Curiously, it appeared the following facts:
• At the global level (master code) the SNP are without effects.
• At the local level, the SNP affects the master codes only at very short distance (study of about fifty bases).
but especially it appears two remarkable results: master code seems "conservative " and invariant under the effect of the SNP and especially, finally, the following remarkable result: SNPs are located in the very large majority of the cases in areas of the genome which are “POLES” or remarkable “SITES” according to the MASTER CODE theory. In other words, there is a very strong correlation between the site of the SNP and the character “Functional Site" (hollow minima or tops maxima) via the Genomics/Proteomics analysis of the Master Code.
However, although these areas are functionally strategic at very long distance, the effect of the SNP is only, contradictorily, LOCAL only. One thus sought "meaning" to the information of SNPs following our point of view of “Master Code". Chromosome 1 region bands 1q31 1q32.1. The contig has 209216 bps without undetermined. So, various "points of sights” were studied based on this sample contig, meanwhile they are not reported at this time in this draft report... focusing our attention on the following major result:
Conclusion
We will retain here the point most remarkable:
"The SNPs are LOCATED in strategic and functional sites areas within the genome. Beyond the nucleotide value of a SNP (mutation/polymorphism), it is thus much more its address which is very important ".
Finally, analyzing the SNP locations within genomic contigs establishes the evidence of a strong link between the “functional sites” proposed by master code law and the exact locations of SNPs within the genome. In other hand: SNPs locations are functional.
Acknowledgements
Thanks to my friend Gilles Roquette with whom this adventure of pioneers (1992-1993, 2000-2001) was dubbed genum (numerical genetics). We especially thank also Dr. Robert Friedman M.D. practiced nutritional and preventive medicine in Santa Fe, New Mexico, we also thank the mathematician Pr. Diego Lucio Rapoport (Buenos aires), and Pr. Luc Montagnier, medicine Nobel prizewinner for their interest in my research of biomathematical laws of genomes.