Research Article - (2025) Volume 11, Issue 5
Stimulation of Transforming Growth Factor-Β3 Signaling by Compressive Force in MC3T3-E1 Cells
Moeko Togawa1,
Akira Nakajima1,2*,
Nichika Fukumashi2,
Hitoshi Kariya2,
Kotoe Mayahara1,2,
Takayuki Kawato3 and
Mitsuru Motoyoshi1,2
1Department of Orthodontics, Nihon University School of Dentistry, 1-8-13 Kanda Surugadai, Chiyoda-ku, Tokyo 1018310, Japan
2Department of Dentistry, Nihon University School of Dentistry, 1-8-13 Kanda Surugadai, Chiyoda-ku, Tokyo 1018310, Japan
3Department of Oral Health Sciences, Nihon University School of Dentistry, Tokyo, Japan
*Correspondence:
Akira Nakajima, Department of Dentistry, Nihon University School of Dentistry, 1-8-13 Kanda Surugadai, Chiyoda-ku, Tokyo 1018310,
Japan,
Email:
Received: 23-Jun-2024, Manuscript No. IPBMBJ-24-20490;
Editor assigned: 26-Jun-2024, Pre QC No. IPBMBJ-24-20490 (PQ);
Reviewed: 10-Jul-2025, QC No. IPBMBJ-24-20490;
Revised: 05-Feb-2025, Manuscript No. IPBMBJ-24-20490 (R);
Published:
12-Feb-2025
Abstract
Objective: Expression of transforming growth factor (TGF)-β is strongly associated with osteoblast differentiation. However, the role of TGF-β3 in mediating the effects of compressive force as a mechanical stress on osteoblasts remains unclear. Here, we examined the effects of compressive force on the expression of TGF-β3, and downstream signaling pathways comprising inflammatory cytokines in osteoblasts. Effects of siRNA-mediated knockdown of TGF-β3 on Smad-dependent and Smadindependent signaling pathways, and on inflammatory cytokines were also examined.
Design: Cultured MC3T3-E1 osteoblast-like cells were subjected to a continuous compressive force (0.5, 1.0, or 2.0 g/cm2) for 30 min, 1 h, and 3 h. TGF-β3 expression was examined using real-time polymerase chain reaction and western blot analysis. Total/phosphorylation levels of Smad2, Smad3, ERK1/2, and p38 were determined using western blot analysis. Cox2 and IL-6 levels were also measured using western blot analysis and real-time polymerase chain reaction.
Results: The mRNA and protein levels of TGF-β3 were significantly increased upon application of 1.0 g/cm2, but not 0.5 and 2.0 g/cm2, compressive force for 1 h, relative to the respective levels in untreated control cells. At 1.0 g/cm2, compressive force also increased the phosphorylation of Smad2, Smad3, ERK1/2, and p-38, and the expressions of COX-2 and IL-6. The increased expression was attenuated by pretreatment with siRNA against TGF-β3.
Conclusion: The present findings indicate that 1.0 g/cm2 compressive force can induce the expression of inflammatory cytokines via TGF-β3 signaling in osteoblasts. These results could be suggested the critical role of TGF-β signaling in bone formation.
Keywords
TGF-β3 signaling; Bone formation; Smad-independent signaling; MAPK signaling; Compressing force; MC3T3-E1 cells
Introduction
Transforming Growth Factor-β (TGF-β) plays a role in cell proliferation, migration, and apoptosis through processes partially controlled via complex adhesive interactions between cellular receptors [1-3]. TGF-β has three main isoforms: TGF- β1, TGF-β2, and TGF-β3 [4,5]. TGF-β3 is most commonly involved in developmental processes that occur in affected palatal and lung development, such as epithelial-mesenchymal interactions, cell growth, extracellular matrix production, and remodeling [5,6].
Orthodontic tooth movement is caused by bone remodeling. Various molecules are present in periodontal tissue, including alveolar bone osteoblast/osteoclast interactions, during tooth movement. In stimulation of bone formation by mechanical stress such as orthodontic force, cyclic mechanical strain results in a significant increase in the levels of active TGF-β1, with no effects on the amount of total TGF-β1. TGF-β1 activation also leads to the phosphorylation of Smad leading to the transcriptional activation of downstream mediators and auto-induction of TGF-β1. In a previous study, we determined the effects of compressive force on the TGF-β1 and TGF-β2 signaling pathways. We discovered that application of 1.0 g/ cm2 compressive force for 3–6 h induced bone-specific transcription factors via the autocrine action of TGF-β1 and TGF-β2 signaling in osteoblasts.
Whether the optimal mechanical stress imposed by compressive force stimulates the specific functional role of TGF-β3 during tooth movement is not clear. Furthermore, whether the Smad-dependent and Smad-independent pathways are involved in the regulatory mechanism is unclear. The mechanism of TGF-β3 stimulation for bone formation during osteoblastic function, which involves a relationship of TGF-β3 with inflammatory cytokines, such as Cyclooxygenase-2 (COX-2) and Interleukin-6 (IL-6), is not clearly identified.
To identify the functional role of TGF-β3 in osteoblastic function via the Smad-dependent and Smad-independent signaling pathways, we stimulated TGF-β3 in the MC3T3-E1 cell line using an optimum compressive force. This study aimed to determine whether compressive forces affect osteogenesis by modulating TGF-β3 expression through the Smad-dependent and independent signaling pathways using a small interfering RNA (siRNA) method.
Materials and Methods
Cell Culture and Application of Compressive Force
MC3T3-E1, a mouse calvarial cell line, was obtained from the RIKEN BioResource Center (Tsukuba, Japan) and used as an osteoblast-like cell line. The cells were maintained in α-minimal essential medium (α-MEM; Gibco BRL, Rockville, MD, USA) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS; HyClone Laboratories, Logan, UT, USA) at 37°C in a humidified atmosphere containing 95% air and 5% CO2.
The cells were seeded in 100 mm cell culture dishes at a density of 2.0 × 104 cells/cm2 and incubated overnight until confluent. They were then compressed continuously by application of a uniform compression method, as described previously. Briefly, thin round glass plates were placed over a layer of confluent cells without FBS, and compressive force was adjusted by placing a lead weight on the glass plates. Confluent cells in a culture dish (inner diameter, 83 mm; thickness, 1.0 mm) were covered with the glass plate (inner diameter, 78 mm; thickness, 0.85 mm) (Figure 1). The weight was positioned such that the force was evenly distributed across the cell monolayer.
The cells were subjected to 0.5, 1.0, or 2.0 g/cm2 compressive force for 30 min, 1 h, and 3 h. Control cells were covered with the same type of glass plates, but without lead weights, which produced a compressive force of 0.035 g/cm2. The possibility that the removal of the glass plates might cause static electric charge, which could affect the cells, was controlled by using glass plates on both the compressive force-treated and control cells (Figure 1).
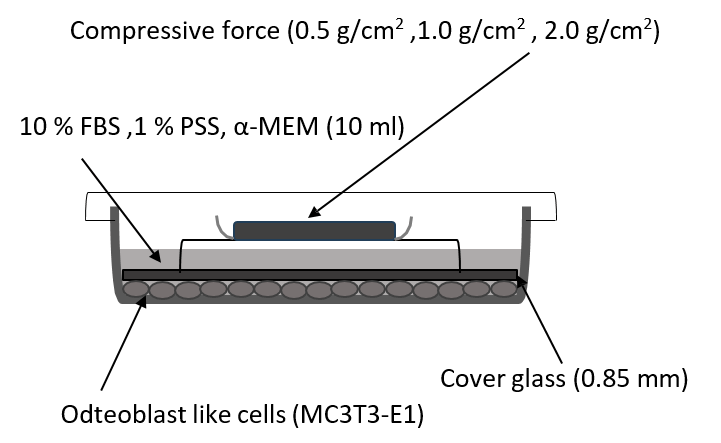
Figure 1: Diagrammatic representation of the application of compressive force.
Note: Confluent cells in a culture dish (inner diameter, 83 mm; thickness, 1.0 mm) were covered with a glass plate (inner diameter, 78 mm; thickness, 0.85 mm). The weight was positioned such that the force was evenly distributed across the cell monolayer.
siTGF-β3 Transfection
To identify the effect of compressive force on TGF-β function, MC3T3-E1 cells were seeded in 100 mm cell culture dishes at a density of 2.0 × 104 cells/cm2. Experimental groups were treated with siRNA specific for TGF-β3 (Thermo Fisher Scientific, Silencer Select siRNA; Assay ID 186708, 186709) before stimulating with the compressive force. The sequences of TGF-β3 siRNA pairs were 5′-UUA CCA AUU UGG UCA CUG UCA UGGA-3′ and 5′-UCC AUG ACA GUG ACC AAA UUG GUAA-3′ (Invitrogen Japan, Stealth). The stimulation was examined for up to 1 h with or without compressive force.
For mRNA assessment, the cell samples received either siTGF- β3 or nontargeting control siRNA (siControl; Stealth RNAi™ siRNA Negative Control Lo GC Duplexes, Invitrogen Japan) at a final concentration of 25 pM. siRNA complexes were formed using RNAiMAX (Invitrogen) in Opti-MEM (Gibco). For all the conditions, antibiotics were omitted, as recommended by the manufacturer, to preserve cell viability during the transfection process.
Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total mRNA was isolated from cultured MC3T3-E1 cells using a commercially available kit (NucleoSpin RNA; TaKaRa, Tokyo, Japan). Aliquots containing equal amounts of mRNA were subjected to real-time RT-PCR. First-strand cDNA synthesis was carried out using 1 μg of DNase-treated total mRNA in 20 μL of a solution containing first-strand buffer, 50 ng random primers, 10 mM dNTP mixture, 1 mM DTT, and 0.5 U reverse transcriptase at 42°C for 60 min. The cDNA mixtures were diluted five-fold using sterile distilled water, and 2 μL aliquots were subjected to real-time RT-PCR using SYBR Green I dye. Real-time RT-PCR was performed in a final reaction volume of 25 μL containing 1X R-PR buffer, 1.5 mM dNTP mixture, 1X SYBR Green I, 15 mM MgCl2, 0.25 U ExTaq polymerase realtime RT-PCR version (TaKaRa, Tokyo Japan), and 20 mM specific primers (TaKaRa, Tokyo Japan); the sequences of primers are listed in Supplementary Table 1.
PCR was performed on a thermal cycler (Smart Cycler, Cepheid, Sunnyvale, CA) and data were analyzed using the Smart Cycler software (ver. 1.2d). The cycling conditions were as follows: 95°C for 3 s and 68°C for 20 s for 35 cycles. Measurements were performed at the end of the annealing step at 68°C in each cycle. The specificity of the RT-PCR products was verified by adding melting curve analysis between 68 and 94°C. All real-time RT-PCR runs were performed in triplicate, and the mRNA expression levels were calculated and normalized against GAPDH mRNA levels.
Western Blot Analysis
To obtain whole-cell extracts, MC3T3-E1 cells were cultured with or without compressive force stimulation, rinsed with phosphate-buffered saline, and then lysed by sonicating three times (10 s each time) in a buffer comprising 50 mM Tris–HCl, 0.1% Triton X-100, 0.1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride. Aliquots containing equal amounts of protein were subjected to Sodium Dodecyl Sulfate- Polyacrylamide Gel Electrophoresis (SDS-PAGE). Samples were electrophoresed on 10–12% polyacrylamide gels and transferred onto polyvinylidene fluoride membranes using a semidry transfer unit. The 1.0% blocking solution (Roche, Basel, Switzerland; #11096176001) in Tres-buffered saline solution (pH 7.6) were performed for blocking and the antibodies diluted by 0.5 % blocking solution in Tris-buffered saline solution.
Using SNAP i.d. 2.0 (Millipore Sigma, Massachusetts, United States), the membranes were probed with anti-TGF-β3, antitotal- Smad2, anti-phosphor Smad2, total-Smad3, anti-phosphor Smad3 (Cell Signaling Technology Japan, Tokyo, Japan; dilution 1:500, polyclonal rabbit antibodies) to examine the involvement of the Smad-dependent mechanism, and with anti-total- ERK1/2, anti-phospho-ERK1/2, total-p38, anti-phosho-p38 (Cell Signaling Technology Japan, Tokyo, Japan; dilution 1:500, polyclonal rabbit antibodies) as Smad-independence signaling. And COX-2, IL-6 as inflammatory cytokines signaling (Thermo Fisher Scientific K.K., Japan; dilution 1:500, polyclonal rabbit antibodies). The anti-GAPDH antibodies as an internal standard (Millipore, MA, USA; monoclonal mouse antibody, dilution 1:500) followed by a biotin-conjugated secondary antibody (Invitrogen, CA, USA; dilution 1:10,000) (Supplementary Table 2). The membranes were then treated with horseradish peroxidase-conjugated streptavidin. Immunoreactive proteins were visualized using a chemiluminescence kit (Amersham Life Science, Buckinghamshire, UK), according to the manufacturer’s instructions. The band intensities of the immunoblots were quantified using a computer scanner (Epson EP-884AB, Seiko Epson, Tokyo, Japan) and digital image analysis software (ImageJ, version 1.54d, Wayne Rasband and contributors NIH, USA) [7].
Statistical Analysis
All the experiments were performed at least five independent times. The non-parametric procedures of the Wilcoxon signed-rank test were conducted for statistical analysis to compare between each experimental group.
The data are expressed as the mean ± standard error of the mean, where n represents the number of experimental palates. Statistical analyses were performed using SPSS software (IBM Corporation, Armonk, NY, USA). Statistical significance was defined as a P value of less than 0.05 (P<0.05).
Results
Effect of Compressive Force on the expression of TGF-β3
Real-time RT-PCR of unstimulated MC3T3-E1 cells or cells stimulated with a continuous compressive force (0.5, 1.0, and 2.0 g/cm2) indicated that the expression of TGF-β3 mRNA increased gradually, peaking at 1 h, and then decreased gradually (Figure 2A). The expression of TGF-β3 mRNA was increased in the 1.0 g/cm2 compressive force group compared with that in the control, and 0.5 and 2.0 g/cm2 compressive force groups at 1 h (P<0.05, n=5; Figure 2A) [<a href="#8" title="8">8</a>,<a href="#9" title="9">9</a>].
Western blot analysis confirmed that among cells stimulated with 0.5, 1.0, and 2.0 g/cm2 compressive force for 1 h, the expression of TGF-β3 protein was significantly higher for those treated with 1.0 g/cm2 compressive force than for cells under any other condition (P<0.05, n=5; Figure 2B).
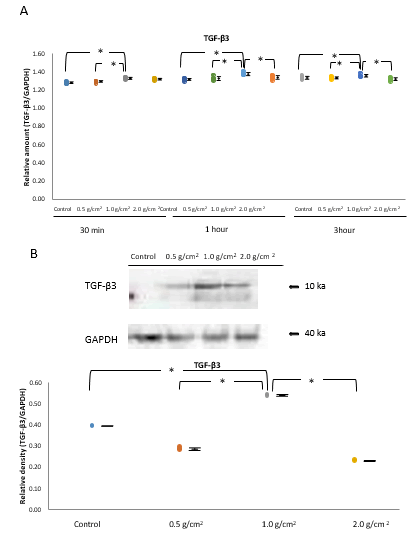
Figure 2: Effects of compressive force on the expression of TGF-β3.
Note: A) The expression of TGF-β3 mRNA increased gradually, peaked at 1 h, and decreased gradually, thereafter. The expression of TGF-β3 mRNA was higher in the 1.0 g/cm2 compressive force group than in the control, and 0.5 and 2.0 g/cm2 compressive force groups at 1 h (*P<0.05, n=5). B) Western blot analysis confirmed that in cells stimulated with 0.5, 1.0, and 2.0 g/cm2 for 1 h, the expression levels of TGF-β3 were significantly higher in the 1.0 g/cm2 compressive force group than under any other condition in western blot analysis (*P<0.05, n=5; Wilcoxon signed rank test).
Effects of Compressive Force on the Phosphorylation of Smad2, Smad3, ERK1/2, and p38
Western blot analysis revealed that in the 1.0 g/cm2 compressive force group, the phosphorylation of Smad2 and Smad3 was higher than that in the control, and 0.5 and 2.0 g/ cm2 compressive force groups at the 1 h (P<0.05, n=5; Figure 3A and 3B).
To determine the effects of compressive force on ERK1/26 and p-38, which are components of the Smadindependent signaling pathway, we determined this phosphorylation using western blot analysis. The levels of phosphorylated-ERK1/2 were significantly increased in the 1.0 g/cm2 compressive force group compared with those in the control, and 0.5 and 2.0 g/cm2 compressive fore groups (P<0.05, n=5; Figure 3C). The level of phosphorylated-p38 in the 1.0 g/cm2 compressive force group was significantly higher than in the other examined groups (P<0.05, n=5; Figure 3D).
The total protein expression (Smad2, Smad3, ERK1/2, and p38) levels were approximately equal in all samples, which levels were similar in control and compressive force-treated cells at 1 h (Figure 3 A-D).
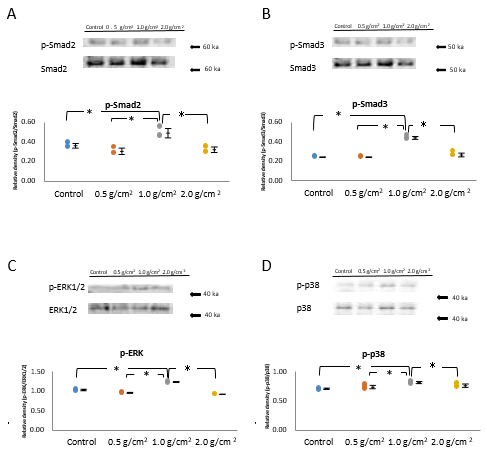
Figure 3: Effects of compressive force on the phosphorylation and total of Smad2, Smad3, ERK1/2, and p38.
Note: (A,B) The 1.0 g/cm2 compressive force group exhibited higher levels of phosphorylated Smad2 and Smad3 than did the control, and 0.5 g/cm2 and 2.0 g/cm2 compressive force groups at 1 h (*P<0.05, n=5; Wilcoxon signed rank test). (C,D) To determine the effects of compressive force on ERK1/26 and p-38, the components of Smad-independent signaling pathway, the levels of phosphorylated of ERK1/2 and p38 were determined using Western blot analysis. C) The GAPDH expression levels were equal in the control and all compressive force groups; however, the levels of phosphorylated-ERK1/2 were significantly increased in the 1.0 g/cm2 compressive force group compared with those in the control, and 0.5 and 2.0 g/cm2 compressive fore groups (*P<0.05, n=5; Wilcoxon signed rank test). D) The phosphorylated-p38 levels were also significantly increased in the 1.0 g/cm2 compressive force group than other examined groups (*P<0.05, n=5; Wilcoxon signed rank test).
Effects of Compressive Force on the Expression of COX-2 and IL-6
The evaluation of the expression of inflammatory cytokines and osteogenic transcription factors in osteoblasts using realtime RT-PCR analysis indicated that the 1.0 g/cm2 compressive force group had significantly higher mRNA levels of inflammatory cytokines COX-2and IL-6 (P<0.05, n=5; Figure 4A-a and 4B-a) than those in the control, and 0.5 and 2.0 g/cm2 compressive force groups at 1-h, correlating with TGF-β3 expression.
The protein expression using the western blot were also obtained that the 1.0 g/cm2 compressive force group detected significantly the highest protein levels of COX-2 and IL-6 as inflammatory cytokines (P<0.05, n=5; Figure. 4A-b and 4B-b).
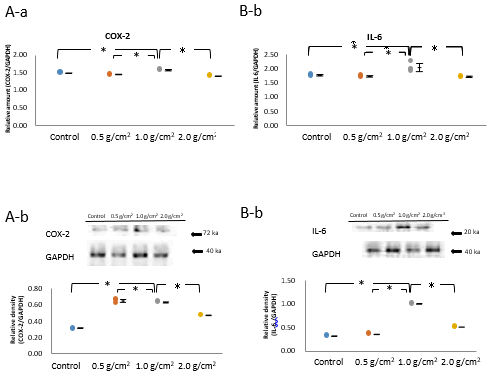
Figure 4: Effects of compressive force on the mRNA and protein expression of COX-2 and IL-6.
Note: A-a, b, B-a, b) The 1.0 g/cm2 compressive force group exhibited significantly higher mRNA levels of the inflammatory cytokines, COX-2 and IL-6 (*P<0.05, n=5) than did the control, and 0.5 and 2.0 g/cm2 compressive force groups at 1 h, the time point at which the stimulation of TGF-β3 expression by compressive force was maximum (*P<0.05, n=5; Wilcoxon signed rank test). C-a, b) Real-time RT-PCR and Western blot of samples at the 1 h time point showed significant increase in the mRNA expression of Runx2, an osteogenic transcription factor, in the 1.0 g/cm2 compressive force group compared with that in the control, and 0.5 and 2.0 g/cm2 compressive force groups (*P<0.05, n=5; Wilcoxon signed rank test). D-a, b) The both Western blot and real-time PCR also indicated that the 1.0 g/cm2 compressive force group had significantly increased mRNA expression of the osteogenic transcription factor Dlx5 (*P<0.05, n=5; Wilcoxon signed rank test), compared with that in the control, and 0.5 and 2.0 g/cm2 compressive force groups at 1 h (*P<0.05, n=5; Wilcoxon signed rank test).
Effect of siTGF-β3 Transfection on Stimulation with 1.0 g/cm2 Compressive Force at 1 h
The expression of TGF-β3 mRNA in cells subjected to 1.0 g/cm2 compressive force (Control+1.0 g/cm2 group) and in siControltreated cells subjected to 1.0 g/cm2 compressive force (siControl+1.0 g/cm2 group) was significantly higher than in control cells not subjected to compressive force (Control group) (P<0.05, n=5; Figure 5A). The TGF-β3 mRNA level in siTGF-β3-treated cells subjected to 1.0 g/cm2 compressive force cell (siTGF-β3+1.0 g/cm2 group) was significantly lower than that in Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups (P<0.05, n=5; Figure 5A), being similar to that in the control group.
The levels of TGF-β3 protein in Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups were higher than those in the Control group. The increase in expression was significantly reduced by siTGF-β3 treatment (P<0.05, n=5; Figure 5B).
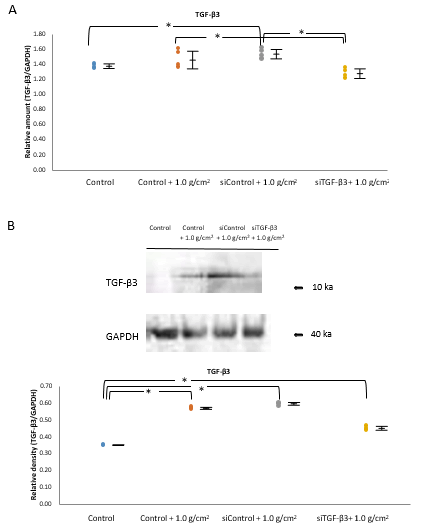
Figure 5: Effect of siTGF-β3 transfection on the stimulation of TGF-β3 expression by 1.0 g/cm2 compressive force at 1 h.
Note: A) The expression of TGF-β3 mRNA in cells subjected to 1.0 g/cm2 compressive force (Control+1.0 g/cm2 group) and in siControl-treated cells subjected to 1.0 g/cm2 compressive force (siControl+1.0 g/cm2 group) was increased significantly compared with that in cells not subjected to compressive force (Control group) (*P<0.05, n=5; Wilcoxon signed rank test). The expression of TGF-β3 mRNA in siTGF-β3-treated cells subjected to 1.0 g/cm2 compressive force (siTGF-β3+1.0 g/cm2 group) was significantly deceased compared with that in the Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups, which was approximately same level to that in the Control group (*P<0.05, n=5; Wilcoxon signed rank test). (B) The expression levels of TGF-β3 in the Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups was increased compared with that in the Control group. The increase in expression caused by compressive force was significantly reduced by siTGF-β3 treatment in Western blot analysis (*P<0.05, n=5; Wilcoxon signed rank test).
Effect of siTGF-β3 transfection on compressive force-induced phosphorylation of proteins in the Smad-dependent and Smad-independent signaling pathways
The relative levels of phosphorylated Smad2 and Smad3 in Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups were significantly higher than those in the control group (P<0.05, n=5; Figure 6A and 6B). The phosphorylation of Smad2 and Smad3 in the siTGF-β3+1.0 g/cm2 group was significantly lower than that in the control+1.0 g/cm2 and siControl+1.0 g/ cm2 groups, which was indicated similar level to that in the control group (P<0.05, n=5; Figure 6A and 6B).
The relative levels of phosphorylated ERK-1/2 and p38 in control+1.0 g/cm2 and siControl+1.0 g/cm2 groups were significantly higher than in the control group, and there was no difference between the control+1.0 g/cm2 and siControl +1.0 g/cm2 groups. The increase in phosphorylation of ERK1/2 and p38 in the siTGF-β3+1.0 g/cm2 group was significantly decreased compared with that in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups, which was approximately same expression to that in the control group (P<0.05, n=5; Figure 6C and 6D). In contrast, the increase in phosphorylation of ERK1/2 and p38 by compressive force was significantly decreased via siTGF-β3 treatment to similar levels to those in the control group (P<0.05, n=5; Figure 6C and 6D).
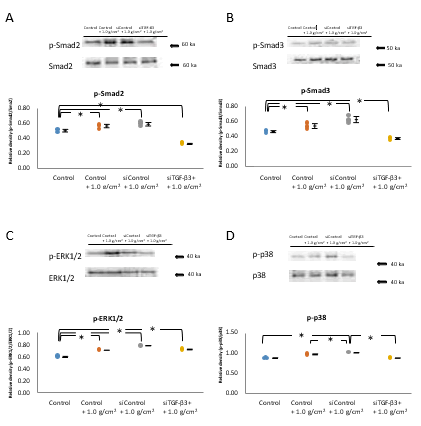
Figure 6: Effect of siTGF-β3 transfection on compressive forceinduced phosphorylation of proteins in Smad-dependent and Smad-independent pathways.
Note: A, B) The levels of phosphorylated Smad2 and Smad3 in cells subjected to 1.0 g/cm2 compressive force (Control+1.0 g/ cm2 group) and in siControl-treated cells subjected to 1.0 g/ cm2 compressive force (siControl+1.0 g/cm2 group) were increased significantly compared with that in cells not subjected to compressive force (Control group) (*P<0.05, n=5; Wilcoxon signed rank test). The phosphorylation of Smad2 and Smad3 in siTGF-β3-treated cells subjected to 1.0 g/cm2 compressive force (siTGF-β3+1.0 g/cm2 group) was significantly deceased compared with that in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups, and was similar to that in the control group (*P<0.05, n=5; Wilcoxon signed rank test). The phosphorylation of ERK-1/2 and p38, proteins in the Smad-independent pathway, in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups was as significantly increased compared with that in the control group, and there was no difference between the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups. C,D) The increase in phosphorylation of ERK1/2 and p38 in the siTGF-β3+1.0 g/cm2 group was significantly decreased compared with that in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups, which was approximately same value to that in the control group (*P<0.05, n=5; Wilcoxon signed rank test). In contrast, the increase in phosphorylation of ERK1/2 and p38 by compressive force was significantly decreased by siTGF-β3 treatment to levels similar to those in the Control group (*P<0.05, n=5; Wilcoxon signed rank test).
Effect of siTGF-β3 Transfection on Compressive Force- Induced Increase in the Expression of Inflammatory Cytokines
The expression of COX-2 in the Control+1.0 g/cm2 and siControl+1.0 g/cm2 groups was increased significantly compared with that in the control group (P<0.05, n=5; Figure 7A-a,b). The increase in the expression of COX-2 by 1.0 g/cm2 compressive force was significantly reduced by siTGF-β3 treatment to a level similar to that in the control group (P<0.05, n=5; Figure 7A-a, b).
The expression of IL-6 in the control+1.0 g/cm2 and siControl +1.0 g/cm2 groups was significantly higher than that in the control group (P<0.05; Figure 7B-a,b). The increase in expression caused by 1.0 g/cm2 compressive force in the control +1.0 g/cm2 and siControl+1.0 g/cm2 groups was significantly reduced by siTGF-β3 treatment (P<0.05, n=5; Figure 7B-a,b).
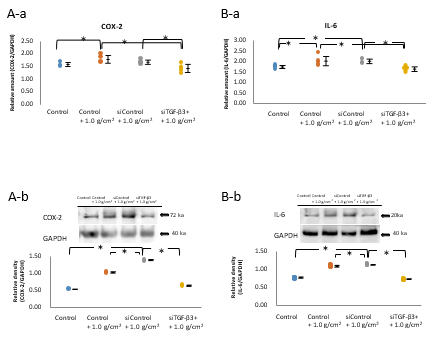
Figure 7: Effect of siTGF-β3 transfection on compressive forceinduced increase in the mRNA and protein expressions of inflammatory cytokines.
Note: A-a, b, B-a, b) The expression of COX-2 in cells subjected to 1.0 g/cm2 compressive force (Control+1.0 g/cm2 group) and in siControl-treated cells subjected to 1.0 g/cm2 compressive force (siControl+1.0 g/cm2 group) was increased significantly compared with that in cells not subjected to compressive force (Control group) (*P<0.05, n=5; Wilcoxon signed rank test). The increase in the expression of COX-2 by 1.0 g/cm2 compressive force was significantly reduced by siTGF-β3 treatment to a level similar to that in the control group (*P<0.05, n=5; Wilcoxon signed rank test). The expression of IL-6 in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups was significantly higher than that in the control (*P<0.05, n=5; Wilcoxon signed-rank test). The increase in expression caused by 1.0 g/cm2 compressive force in the control+1.0 g/cm2 and siControl+1.0 g/cm2 groups was significantly reduced by siTGF-β3 treatment (*P<0.05, n=5; Wilcoxon signed-rank test).
Discussion
The main findings of the present study were that 1.0 g/cm2 compressive force stimulated the expression of TGF-β3 in osteoblast-like MC3T3-E1 cells and also stimulated the phosphorylation of Smad2 and Smad3 in the Smad-dependent signaling pathway, and of ERK1/2 and p38, in the Smadindependent pathway. The expression levels of the inflammatory cytokines, COX-2 and IL-6 were also increased upon application of 1.0 g/cm2 compressive force. These stimulatory effects were not observed for cells subjected to 0.5 or 2.0 g/cm2 compressive force. The stimulatory effects of 1.0 g/cm2 compressive force were significantly reduced by siTGF-β3 treatment of cells, and the expression levels were similar to those in control cells that were not subjected to compressive force. These findings are consistent with those of prior studies showing that 1.0 g/cm2 compressive force is an optimal mechanical stress for facilitating osteogenesis in osteoblasts by increasing the levels of type I collagen, bone sialoprotein, and bone morphogenetic protein, whereas weak or excessive mechanical stress attenuates these processes [<a href="#8" title="8">8</a>,<a href="#9" title="9">9</a>].
The sequence of TGF-β3 is 86% similar to that of TGF-β1 and 91% similar to that of TGF-β2 10. However, despite TGF-β2 and β3 sharing the highest level of sequence similarity among the three isoforms, TGF-β2 binds to the TGF-β receptor II (TβRII) in a manner different from that of TGF-β1 and β3, and involves different residues. Furthermore, although both TGF- β1 and β3 are capable of direct binding to TβRII, presentation of TGF-β2 to the receptor requires the presence of a coreceptor (beta glycan or endoglin), which may explain the differences in activities of TGF-β2 and β3.
TGF-β3 plays an important role in normal developmental biology including those in systems, such as the craniofacial, heart, lung, and breast, and displays isoform-specific biology at both the in vivo and in vitro level. An understanding of the unique biology of TGF-β3 is important to comprehend its role in the osteoblastic mechanism. Differences in the functional role of TGF-β biology have been studied in some depth for the development of the palate. In palatal development, TGF-β3 is strongly expressed in mesial edge epithelia cells before the contact and fusion of opposing palatal shelves. It continues to be strongly expressed during palatal fusion in the midline seam of palatal epithelial cells including mesenchymal cells, which undergo epithelial-mesenchymal transformation. Notably, a complete cleft palate is observed in TGF-β3 null mice, even though the palatal mesenchymal shelves in this model have sufficient length and orientation to allow fusion. Moreover, unlike other null mutants exhibiting a cleft palate, TGF-β3 null mice lack other concomitant craniofacial abnormalities. Compared to other ligands, TGF-β3 is more specialized in its patterning of expression during palatogenesis and in its localization to the palatal epithelium cells and, thus, has the potential to fine-tune the fate of mesial edge epithelial cells toward migration, apoptosis, or transformation.
In our previous study on osteoblast response, in MC3T3-E1 cells stimulated, or not, with continuous compressive force (1.0 and 2.0 g/cm2), the expression of TGF-β1 and TGF-β2 increased gradually up to the 3 or 6 h time point, and decreased gradually, thereafter. The levels of TGF-β1 and TGF- β2 mRNAs increased in the 1.0 g/cm2 compressive force group compared with that in the control and 2.0 g/cm2 compressive force groups at the 3 and 6 h time points, respectively. Western blot analysis confirmed that the cells stimulated with 1.0 g/cm2, but not 2.0 g/cm2, continuous compressive force exhibited higher TGF-β1 and TGF-β2 protein levels than the control cells.
Given that TGF-β3 can have positive regulatory effects on osteoblast differentiation, the imposition of 1.0 g/cm2, but not 0.5 and 2.0 g/cm2, compressive force could be presumed to promote osteoblast differentiation. Osteoblast differentiation and bone formation appear to be influenced by Smadindependent as well as Smad-dependent TGF-β3 signaling pathways. For example, the association of TAK1 and TAK1 binding protein 1, which is induced by TGF-β results in activation of the Mitogen-Activated Protein Kinase (MAPK) kinase 3-p38 MAPK signaling cascade that leads to the induction of type I collagen expression, and TAK1 regulates the steady-state protein levels of these two kinases. We also observed a similar function of TGF-β3 under compressive force treatment. Notably, a recent study demonstrated that following TGF-β3 induction, the Smad and ERK/p38 MAPK pathways converged at Runx2 in the control of mesenchymal precursor cell differentiation. However, the issue of Smad-independent signaling induced by TGF-β, i.e., autocrine/paracrine inflammatory cytokines (IL-6 or COX-2 e.g.) activated by mitogen-activated protein kinase (MAPK) signaling cascade, remains to be addressed (Figure 8).
Bone Morphogenetic Proteins (BMPs), which are members of the TGF-β superfamily and share 32–37% sequence homology with TGF-βs, profoundly affect the osteoblast activity. Following BMP induction, Smad1/5, and MAPK cascades converge at Runx2 to control the differentiation of mesenchymal precursor cells. Compressive force was reported to increase the expression of BMPs and their receptors, phosphorylated-Smad1, osteogenic transcription factor (e. g. Runx2 and Dlx5), and to augment in vitro mineralization; these effects were attenuated by BMP receptor antagonism. Previous findings and the results of the present study indicate that both BMP and TGF-β signaling pathways might be stimulated by compressive force and osteoblast differentiation is induced via Smad-dependent and Smadindependent cascades, and that crosstalk of BMP and TGF-β signaling pathways might be related to these phenomena.
In addition, it is also important whether the effect on actin cytoskeletal regulators by TGF-β3 signaling. According to previous study, to maintain their architecture and function, respond to injuries by activating intricate biochemical and physical mechanisms that regulates intercellular communication crucial in maintaining tissue homeostasis. Coordination of the communication occurs through the activity of different actin cytoskeletal regulators, physically connected to extracellular matrix through integrins, generating a platform of biochemical and biomechanical signaling. In the future plan, these suggestions and mechanism such as autocrine/paracrine, cross-talk with related gene, and actin reaction. However, the present study discovered that siTGF-β3 transfection prevented compressive force-induced phosphorylation of Smad2/3, ERK, p38 and the expression of inflammatory cytokines, which supports the notion that Smad-dependent/-independent signaling can also promote osteoblast differentiation via expression of osteogenesis-related transcription factors (Figure 8).
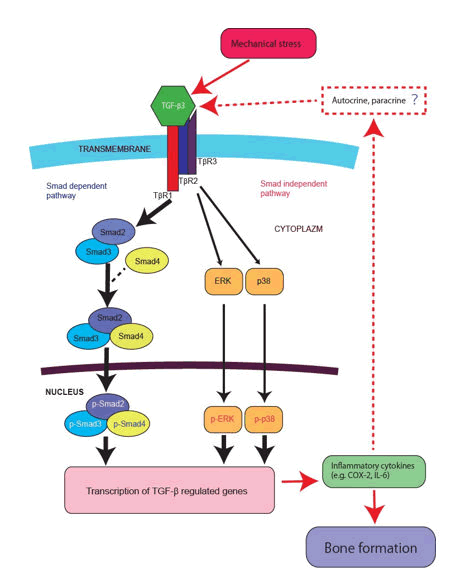
Figure 8: Diagrammatically show TGF-β3 signalling cascade events related with Smad-dependent and -independent with their interactions including inflammatory cytokines by mechanical stress stimulation.
Note: Gene abbreviations are listed below. Transforming growth factor: TGF, TGF receptor: TβR, Cyclooxygenase-2: COX-2, Interleukin 6: IL-6, Extracellular signal-regulated kinases: ERK, p38 MAP Kinase: p38.
Conclusion
In summary, the 1.0 g/cm2 compressive force stimulated the expression of TGF-β3 signaling pathway components, including inflammatory cytokines and osteogenic transcription factors in MC3T3-E1 cells. These osteoblastic differentiation phenomena mediated by TGF-β3 expression were reflected in the expression of both Smad-dependent and Smadindependent signaling pathway molecules, suggesting that optimal force stimulates osteogenic bone formation, at least under the experimental conditions used in the present study. Moreover, siTGF-β3 transfection attenuated the effect of compressive force on phosphorylation of the Smad-dependent and Smad-independent signaling composition. Increased expression of COX-2 and IL-6 induced by compressive force was reduced upon siTGF-β3 transfection in osteoblast cells. The stimulation of the expression of TGF-β3 and signaling pathway proteins by 1.0 g/cm2 compressive force, which sequentially peaked at 1 h, earlier than peaking of the expression of TGF-β2 and TGF-β1.
Conflict Of Cometing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Copyright Transfear Form
The data underlying this article will be shared on reasonable request to the corresponding author
Acknowledgements
The authors thank Professor Yoshio Nakano for the suggestions of statistical analysis, and our colleagues at the department of orthodontics, the department of biochemistry, and the department of oral health sciences, Nihon University School of Dentistry, Japan for their continuous support.
Credit Authorship Contribution Statement
Moeko Togawa: Writing-review and editing, writing-original draft, methodology, investigation, formal analysis, data curation, conceptualization. Akira Nakajima: Writing-review and editing, writing-original draft, methodology, investigation, formal analysis, data curation, conceptualization. Nichika Fukumashi: Writing-review and editing, writing-original draft, methodology, investigation, data curation, conceptualization. Hitoshi Kariya: Writing-review and editing, writing-original draft, methodology, investigation, data curation, conceptualization. Kotoe Mayahara: Writing-review and editing, writing-original draft, methodology, investigation, data curation, conceptualization. Takayuki Kawato: Writing-review and editing, writing-original draft, methodology, investigation, formal analysis, data curation, conceptualization. Mitsuru Motoyoshi: Writing-review and editing, writing-original draft, methodology, investigation, formal analysis, data curation, conceptualization.
Declaration of Competing Interests
None.
Funding
This work was supported by JSPS KAKENHI (Grant Number: JP22K10280) and the Dental Research Center in University School of Dentistry, Graduate School of Dentistry (B).
References
- Nakajima A, F. Shuler C, Gulka AO, Hanai JI (2018) TGF-beta Signaling and the Epithelial-Mesenchymal Transition during Palatal Fusion. Int J Mol Sci. 19(11):3638. [Crossref]
[Google Scholar] [PubMed]
- Peters AS, Brunner G, Krieg T, Eckes B (2015) Cyclic mechanical strain induces TGF-beta1-signalling in dermal fibroblasts embedded in a 3D collagen lattice. Arch Dermatol Res. 307(2):191-197.
[Crossref] [Google Scholar] [PubMed]
- Manokawinchoke J, Limjeerajarus N, Limjeerajarus C, Sastravaha P, Everts V, et al. (2015) Mechanical Force-induced TGFB1 Increases Expression of SOST/POSTN by hPDL Cells. J Dent Res. 94(7):983-989.
[Crossref] [Google Scholar] [PubMed]
- Grafe I, Alexander S, Peterson JR, Snider TN, Levi B, et al. (2018) TGF-beta Family Signaling in Mesenchymal Differentiation. Cold Spring Harb Perspect Biol. 10(5):a022202.
[Crossref] [Google Scholar] [PubMed]
- Wu M, Chen G, Li YP (2016) TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 4:16009.
[Crossref] [Google Scholar] [PubMed]
- Takeyama K, Chatani M, Inohaya K, Kudo A (2016) TGFbeta-2 signaling is essential for osteoblast migration and differentiation during fracture healing in medaka fish. Bone. 86:68-78.
[Crossref] [Google Scholar] [PubMed]
- Nakajima A, Ito Y, Tanaka E, Sano R, Karasawa Y, et al. (2014) Functional role of TGF-beta receptors during palatal fusion in vitro. Arch Oral Biol. 59(11):1192-1204.
[Crossref] [Google Scholar] [PubMed]
- Mitsui N, Suzuki N, Maeno M, Mayahara K, Yanagisawa M, et al. (2005) Optimal compressive force induces bone formation via increasing bone sialoprotein and prostaglandin E(2) production appropriately. Life Sci. 77(25):3168-3182.
[Crossref] [Google Scholar] [PubMed]
- Mitsui N, Suzuki N, Maeno M, Yanagisawa M, Koyama Y, et al. (2006) Optimal compressive force induces bone formation via increasing bone morphogenetic proteins production and decreasing their antagonists production by Saos-2 cells. Life Sci. 78:2697-2706.
[Crossref] [Google Scholar] [PubMed]
Citation: Togawa M, Nakajima A, Fukumashi N, Kariya H, Mayahara K (2025) Stimulation of Transforming Growth Factor-?3
Signaling by Compressive Force in MC3T3-E1 Cells. Biochem Mol Biol. 11:49.
Copyright: © 2025 Togawa M, et al. This is an open-access article distributed under the terms of the Creative
Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the
original author and source are credited.









