- (2010) Volume 11, Issue 1
Emma L Edghill1, Jayne AL Minton1, Christopher J Groves3, Sarah E Flanagan1, Ann-Marie Patch1, Oscar Rubio-Cabezas1,4,5, Maggie Shepherd2, Sigurd Lenzen6, Mark I McCarthy3, Sian Ellard1 and Andrew T Hattersley1
Institutes of 1Biomedical and Clinical Science and 2Health and Social Care, Peninsula Medical
School. Exeter, United Kingdom.
3Oxford Centre for Diabetes, Endocrinology and Metabolism,
University of Oxford. Oxford, United Kingdom.
4Department of Endocrinology, Hospital Infantil
Universitario Niño Jesús;
5Department of Pediatrics, Universidad Autónoma de Madrid. Madrid
Spain.
6Institute of Clinical Biochemistry, Medizinische Hochschule. Hannover, Germany
Received June 29th, 2009 - Accepted October 13th, 2009
Context Approximately 39% of cases with permanent neonatal diabetes (PNDM) and about 11% with maturity onset diabetes of the young (MODY) have an unknown genetic aetiology. Many of the known genes causing MODY and PNDM were identified as being critical for beta cell function before their identification as a cause of monogenic diabetes. Objective We used nominations from the EU beta cell consortium EURODIA project partners to guide gene candidacy. Subjects Seventeen cases with permanent neonatal diabetes and 8 cases with maturity onset diabetes of the young. Main outcome measures The beta cell experts within the EURODIA consortium were asked to nominate 3 “gold”, 3 “silver” and 4 “bronze” genes based on biological or genetic grounds. We sequenced twelve candidate genes from the list based on evidence for candidacy. Results Sequencing ISL1, LMX1A, MAFA, NGN3, NKX2.2, NKX6.1, PAX4, PAX6, SOX2, SREBF1, SYT9 and UCP2 did not identify any pathogenic mutations. Conclusion Further work is needed to identify novel causes of permanent neonatal diabetes and maturity onset diabetes of the young utilising genetic approaches as well as further candidate genes.
Diabetes Mellitus, Type 2; Genetics, Insulin-Secreting Cells
MODY: maturity onset diabetes of the young; OMIM: Online Mendelian Inheritance in Man; PNDM: permanent neonatal diabetes
Monogenic diabetes is a heterogeneous group of disorders characterised most often by pancreatic beta cell dysfunction [1]. Two subtypes of monogenic diabetes include permanent neonatal diabetes (PNDM) and maturity onset diabetes of the young (MODY) [2]. Patients with PNDM present in the first 6 months of life and are often sporadic, whilst MODY presents within families with a strong history of diabetes and at least one subject diagnosed before the age of 25 years. To date there are ten genetic subtypes for PNDM; KCNJ11, ABCC8, INS, GCK, IPF1, FOXP3, EIF2AK3, PTF1A, SLC19A2 and GLIS3 [2]. In MODY there are ten causal genes; HNF1A, GCK, HNF4A, IPF1, HNF1B, NEUROD1, CEL, KLF11, PAX4 and INS [2]. There remains about 37% of cases with PNDM and about 11% with MODY with an unknown genetic aetiology [3]. Many of the known monogenic diabetes genes were identified as biological candidates involved in beta cell development, maintenance and function, such as GCK, KCNJ11, IPF1 and PTF1A [4, 5, 6, 7, 8]. There are many other genes involved in pancreatic development or insulin secretion demonstrated by in vitro studies, animal models, as well as recent type 2 diabetes mellitus genetic studies [9, 10]. It is reasonable to hypothesise that genes for other proteins that are critical for beta cell function could be involved in the pathogenesis of monogenic diabetes. We aimed to sequence candidate genes, selected by international experts in the functioning of beta cells (EURODIA consortium; https://www.eurodia.info/), in patients with monogenic diabetes in whom the major known causes had been excluded.
Subjects
Seventeen cases with PNDM were selected if insulin treated at time of referral and diagnosed at less than 6 months of life. The median age of diabetes onset for the cohort was 8 weeks (range: 0-26 weeks), the median birth weight was 2.83 kg (range: 0.97-3.68) born at a median delivery of 37.5 weeks (range: 36-40 weeks). Two subjects had an affected father. The median age at time of testing was 11 years (range 5-54 years). Mutations in KCNJ11, ABCC8, INS and GCK were excluded.
Eight cases with MODY were selected that were diagnosed within 25 years of age, non-obese and at least two generations affected with diabetes. The median age of onset was 20 years (range: 15-24 years), the median BMI was 24 kg/m2, (range: 23-30 kg/m2) three subjects had an affected mother and five had an affected father. Mutations and deletions in the HNF1A, HNF4A and GCK genes had been excluded.
Candidate Gene Selection
The beta cell experts within the European beta cell consortium EURODIA were asked to nominate 3 “gold”, 3 “silver” and 4 “bronze” genes based on biological or genetic grounds. There were 107 genes (40 gold, 35 silver, 32 bronze) nominated with surprisingly little overlap in the suggestions made, with only one gene attracting three nominations (UCP2). We chose twelve genes from the list to investigate in monogenic diabetes, based on evidence for candidacy. Eight were from the “gold” category (MAFA, NGN3, NKX2.2, PAX4, PAX6, SREBF1, SYT9, UCP2), one from “silver” (LMX1A), and three from “bronze” (ISL1, NKX6.1, SOX2).
Molecular Genetics
Genomic DNA was extracted from peripheral leukocytes using standard procedures. Standard PCR and sequencing techniques amplified each of the coding exons of the 12 candidate genes; ISL1, LMX1A, MAFA, NGN3, NKX2.2, NKX6.1, PAX4, PAX6, SOX2, SREBF1, SYT9 and UCP2 (primers available on request). Changes in the sequence were checked against published polymorphisms. Variants identified were tested for co-segregation or sequenced in control chromosomes.
This work was done with informed consent from the participants on a study protocol which conforms to the ethical guidelines of the declaration of Helsinki and was approved by the local ethics committee.
Frequency, median and range were evaluated as descriptive statistics.
We sequenced twelve candidate genes in 25 cases with PNDM or MODY. No pathological variants were identified.
We did identify novel rare variants in these candidate genes (Table 1). A total of 5 novel non-synonymous variants were identified in PAX4, SYT9, LMX1A, MAFA and SREBF1 genes. None of these 5 variants co-segregated with diabetes within the immediate family (Table 1). In 4 cases the variant was inherited from an unaffected parent. In one patient with PNDM we identified a novel heterozygous G132A variant in LMX1A, this variant was not present in the affected father, diagnosed at 13 years, but the mother was unavailable for testing so a spontaneous mutation cannot be excluded. We also identified 13 previously described common polymorphisms (Table 1).
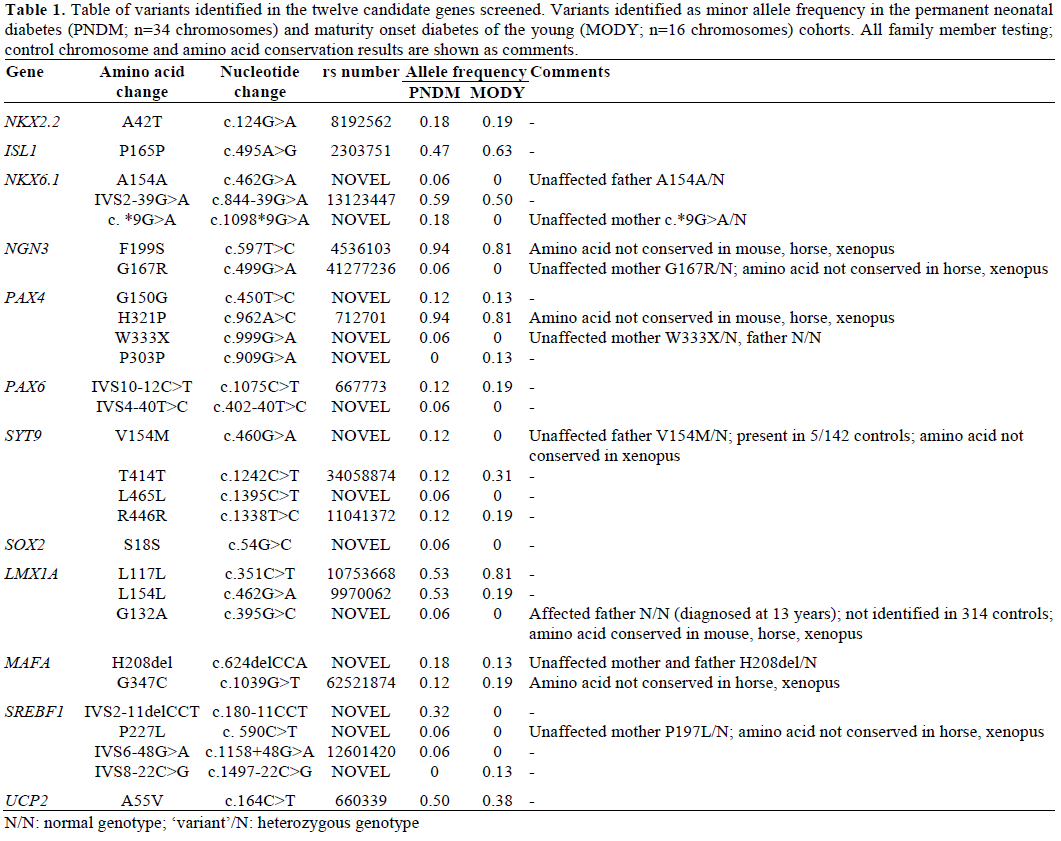
We failed to find mutations in twelve key beta cell genes in patients likely to have monogenic diabetes in whom a genetic aetiology had not been identified. To identify the strongest candidate genes for beta cell function we asked for nominations by internationally acknowledged experts in beta cell function who were partners in the European sponsored EURODIA project. The twelve candidate genes we investigated had sufficient biological and, for some, prior genetic evidence for candidacy. For example the transcription factor genes ngn3, nkx2.2, pax4, pax6, isl1, and mafa are critical for endocrine cell development in rodents [11, 12, 13, 14, 15, 16]. Null mouse models of nkx2.2, ngn3, and mafa result in neonatal hyperglycaemia and in some premature death in the neonatal period [12, 17, 18]. In addition to our study, Nocerino et al. [19] has recently shown that mutations in NGN3 are not a common cause of isolated diabetes, however we have since identified that recessive NGN3 mutations can cause neonatal diabetes and severe diarrhea [20]. Whilst nkx6.1, sox2, lmx1a, syt9 and srebf1 are crucial in maintaining adult beta cell function, insulin release and insulin activity [21, 22, 23, 24, 25]. In man heterozygous SOX2 mutations cause the microphthalmia and esophageal atresia syndrome (Online Mendelian Inheritance in Man; OMIM #206900; https://www.ncbi.nlm.nih.gov/omim), however these subjects have no reported beta cell dysfunction. Moreover, common variation close to PAX4, NGN3 and SREBF1 are associated with type 2 diabetes [26, 27, 28, 29, 30], whilst monogenic mutations in PAX6 result in aniridia and early onset diabetes (OMIM #106210).
We have shown that mutations in these genes are not a common cause of monogenic diabetes in our cohorts. Our sample size means we cannot exclude mutations in these genes being rare causes of PNDM or MODY and these genes remain good candidates for these conditions. We investigated patients with isolated diabetes; therefore some of these genes may be causal in patients with diabetes and other features. The screening strategy used was not designed to detect all types of mutations: the presence of large genomic deletions, mutations in deep intronic or regulatory regions cannot be excluded.
There remains a significant percentage of cases with PNDM or MODY with an unknown genetic aetiology.
There are many more biological candidates involved in pancreatic development, maintenance and function. History will show if the best strategy is to continue with a candidate gene approach or to rely on a genetic approach. There are limited numbers of PNDM or MODY families with multiple affected subjects who do not have one of the known genetic aetiologies. Ultimately novel sequencing approaches may allow the sequencing of large number of genes in a single study.
In conclusion we have shown that a selection of twelve candidate genes, nominated by experts in beta cell function, are not a common cause of isolated monogenic diabetes. Thus other genes, without a presently evident prominent role in beta cell development or maintenance may cause the diabetic metabolic state in these patients.
The authors would like to thank the EURODIA partners for nominating candidate genes and Chris Boustred, Andrew Parrish, and Amna Khamis for technical support. This work was funded by the European Union (Integrated Project EURODIA LSHM-CT-2006-518153 in the Framework Programme 6 of the European-Community). ATH is a Wellcome Trust Research leave fellow.