Review Article - (2017) Volume 3, Issue 1
Theo deVos1* and Bela, Molnar2
1Epigenomics, Inc. Suite 400, 1455 Leary Way NW, Seattle, WA 98107, USA
2Department of Internal Medicine, Semmelweis University, Budapest, Hungary
*Corresponding Author:
Theo deVos
Epigenomics, Inc Suite 400, 1455 Leary Way NW
Seattle, WA 98107, USA.
Tel: 206-883-2916
Fax: 240-747-7052
E-mail: theo.devos@epigenomics.com
Received date: February 21, 2017; Accepted date: February 23, 2017; Published date: February 28, 2017
Citation: deVos T, Molnar B. Screening for Colorectal Cancer Based on the Promoter Methylation Status of the Septin 9 Gene in Plasma Cell Free DNA. J Clin Epigenet. 2017, 3:1. doi: 10.21767/2472-1158.100040
Copyright: © 2017 deVos T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Screening for colorectal cancer is widely considered a cost effective intervention with strong evidence supporting mortality reduction in the screened population. Despite this, global screening rates remain low, even in developed countries. Blood based screening tests have the potential to overcome resistance to screening, thereby increasing overall participation, and saving lives.
Here we review the development of a recently US FDA approved blood based epigenetic screening test for colorectal cancer. The Epi proColon test is based on the Septin 9 (SEPT9) gene promoter methylation status measured in cell free DNA from plasma. While the assessment of DNA methylation has been practiced in laboratories for some time, the approved test provides a standardized and kitted method for isolation of cell free DNA from plasma, reagents and methods for bisulfite conversion and purification of converted DNA (bisDNA) in preparation for DNA methylation analysis by real time PCR. This enables broad dissemination of DNA methylation based testing to laboratories approved to perform standard molecular diagnostics.
In clinical trials, the approved Epi proColon test had sensitivity for colorectal cancer of 68-72%, comparable to commonly used stool tests, at a specificity of 80-82%. The test was well received, with 99.5% of patients in a participation trial agreeing to testing, and of these, when the test was positive, 67% scheduled colonoscopy. Finally, 59% of patients who had a colonoscopy in this study had a finding requiring polypectomy or biopsy. Based on these data, the test was approved as a screening test in the US, for patients who are otherwise unscreened, a key advance for molecular diagnostics applications in the field of clinical epigenetics.
Keywords
Septin 9 gene promoter; Colorectal cancer; Biopsy; Oncology
Background–Methylated Biomarkers in Cell Free Plasma DNA
The development of blood tests based on analysis of cell free nucleic acids, or so called ‘liquid biopsies’ in the past 10 years represents the confluence of advances in different fields of research through the 1980’s and 1990’s. Germane to this journal, rapid advances were being made in the field of epigenetics, particularly in describing modifications of DNA, including DNA methylation and its role in gene regulation [1]. In parallel, a growing body of literature characterized cell free circulating nucleic acids in blood, and the consequent potential for biomarker Screening for Colorectal Cancer Based on the Promoter Methylation Status of the Septin 9 Gene in Plasma Cell Free DNA Received: February 21, 2017; Accepted: February 23, 2017; Published: February 28, 2017 Abstract Screening for colorectal cancer is widely considered a cost effective intervention with strong evidence supporting mortality reduction in the screened population. Despite this, global screening rates remain low, even in developed countries. Blood based screening tests have the potential to overcome resistance to screening, thereby increasing overall participation, and saving lives. Here we review the development of a recently US FDA approved blood based epigenetic screening test for colorectal cancer. The Epi proColon test is based on the Septin 9 (SEPT9) gene promoter methylation status measured in cell free DNA from plasma. While the assessment of DNA methylation has been practiced in laboratories for some time, the approved test provides a standardized and kitted method for isolation of cell free DNA from plasma, reagents and methods for bisulfite conversion and purification of converted DNA (bisDNA) in preparation for DNA methylation analysis by real time PCR. This enables broad dissemination of DNA methylation based testing to laboratories approved to perform standard molecular diagnostics. In clinical trials, the approved Epi proColon test had sensitivity for colorectal cancer of 68-72%, comparable to commonly used stool tests, at a specificity of 80-82%. The test was well received, with 99.5% of patients in a participation trial agreeing to testing, and of these, when the test was positive, 67% scheduled colonoscopy. Finally, 59% of patients who had a colonoscopy in this study had a finding requiring polypectomy or biopsy. Based on these data, the test was approved as a screening test in the US, for patients who are otherwise unscreened, a key advance for molecular diagnostics applications in the field of clinical epigenetics. Keywords: Septin 9 gene promoter; Colorectal cancer; Biopsy; Oncology discovery [2-4]. Along with these developments, technology for the analysis of nucleic acids was rapidly developing, including methods for characterization of modified nucleotides in the genome [5]. Many of these developments were championed in the context of oncology research [6]. This, coupled with the growing sentiment that early detection represented a significant solution to achieve mortality reduction in disease, stimulated both private and academic researchers to identify and develop cancer biomarkers, and in a parallel field, to develop markers for non-invasive prenatal testing [7]. Thus, while a broad array of genetic markers (mutations, re-arrangements) have been identified; analysis of DNA methylation has been an equally fertile area for biomarker discovery and development.
DNA Methylation Status of Gene Sequences as Biomarkers for Disease
Epigenetics research encompasses a growing number of mechanisms for gene regulation mediated through an expanding list of modifications of DNA, RNA and proteins [1]. It should not be surprising therefore, that dysfunction of these regulatory mechanisms, potentially as a consequence of aberrant modifications, can play a significant role in disease and particularly cancer. In this regard, there is a large body of literature describing the global and gene sequence specific status of DNA methylation in oncologic and other disease states [8,9].
The initial observations were that there was an overall reduction in methylation (hypomethylation) in genomic DNA isolated from tumors, compared with healthy tissue [10]. However, it also became apparent that DNA methylation levels varied in the genome, and that the 5’ regions of some genes were CG rich and hypomethylated in normal tissue [11]. As methylation levels were associated with transcription status, hypermethylation of such CpG islands was reported in the transition to cell immortalization [12]. These observations of global hypomethylation and localized hypermethylation have led to multiple rationales for a role for aberrant DNA methylation in cancer. These include an effect on chromosome stability, repression of tumor suppressor genes and derepression of endogenous retroviruses, amongst others. Pertinent to this review, altered DNA methylation patterns are one of the hallmarks of oncogenic transformation, and consequently provide a rich potential source of biomarkers to exploit for molecular diagnostics.
Cell Free DNA in Plasma and Serum
Briefly, the measurement of nucleic acids in blood dates back to the 1930’s, with the first reported measurement of plasma DNA attributed to Mandel and Metais. The measurement of DNA concentrations in the serum or plasma of cancer patients began to appear in the literature in the 1950’s [13-16] with more detailed analyses reported in the 1970’s [17]. The first reference to the potential role for tumor derived plasma DNA having a role in cancer metastases appears as speculation in a 1965 paper describing studies of injection of viral DNA into mice [18]. With the development of improved techniques, evidence for tumor specific DNA in circulation was reported by Stroun et al. in the late 1980’s [19]. Higher sensitivity, sequence specific PCR methods allowed detection of tumor specific mutations, and subsequently, application of bisulfite conversion and methylation specific PCR also allowed for the detection of tumor specific methylation changes in plasma as summarized by Laird [5,20].
Thus, by the late 1990’s cancer specific DNA methylation differences represented a potential source of biomarkers, and the nascent study of cell free nucleic acids in plasma was poised for exponential growth. The general concept for this approach is illustrated in Figure 1.
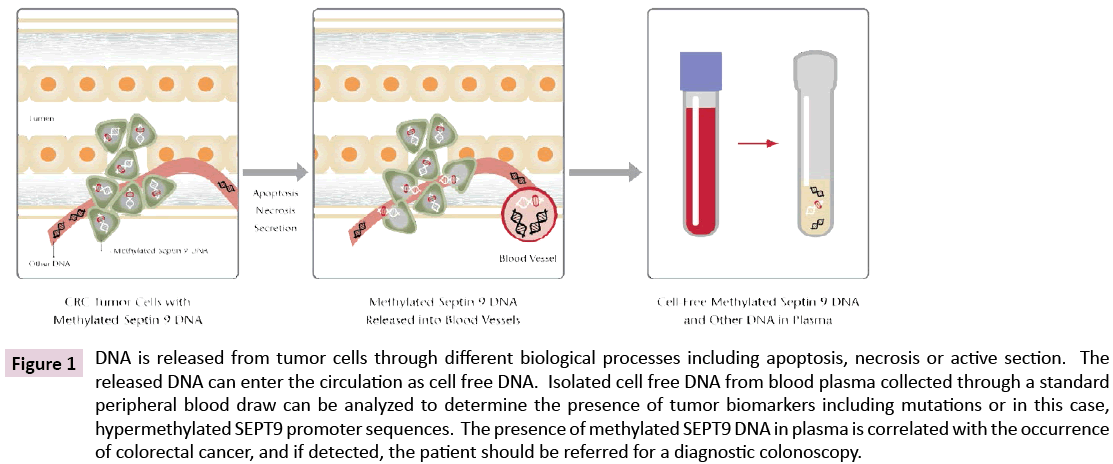
Figure 1: DNA is released from tumor cells through different biological processes including apoptosis, necrosis or active section. The released DNA can enter the circulation as cell free DNA. Isolated cell free DNA from blood plasma collected through a standard peripheral blood draw can be analyzed to determine the presence of tumor biomarkers including mutations or in this case, hypermethylated SEPT9 promoter sequences. The presence of methylated SEPT9 DNA in plasma is correlated with the occurrence of colorectal cancer, and if detected, the patient should be referred for a diagnostic colonoscopy.
SEPT9 Promoter Methylation–Discovery and Development of Methylated SEPT9 and Approval of Epi ProColon
Discovery and characterization
As outlined above, changes in DNA methylation are an early hallmark in the process of oncogenic transformation. Different strategies have been used to identify and characterize these changes, ranging from analysis of specific candidate gene sequences to genome wide approaches. The discovery process to identify methylation markers for colorectal cancer included both, using arbitrarily primed PCR methods as well as methylation hybridization arrays, and methylation specific PCR [21]. It is clear from this and other studies that the challenge in the approach is not finding methylation differences, of which there are hundreds, but in filtering these differences to identify target sequences that have practical value for the intended diagnostic purpose [22].
The objective of these studies was the development of a marker panel to be used as a blood based screening test for colorectal cancer. This objective imposed the marker criteria: 1) Sequences should be differentially methylated in cancer tissue and preferably in adenoma/polyp tissue; 2) Amenable to PCR amplification following bisulfite conversion; 3) Absent in cell free DNA in healthy subjects; 4) Present in detectable quantities in cell free DNA extracted from plasma or serum of patients with colorectal cancer. A collection of several hundred DNA methylation differences discovered in genome wide and microarray studies were selected based on the ability to develop PCR assays, then counter selected based on positivity using peripheral blood leukocyte DNA, and subsequently DNA pools isolated from plasma of healthy subjects, leading to a shortlist of 3 candidate markers [22]. Of these, the promoter methylation status of the SEPT9 gene showed the best correlation with colorectal cancer both in tissue DNA and in cell free DNA isolated from plasma [22,23]. This was supported by laser capture microdissection analysis, showing methylation changes of the SEPT9 promoter associated with tumor progression [24] and in studies of tissue and plasma from the same patients [25]. Though several hundred marker candidates were screened in the discovery process, sensitivity gains from paneling with methylated SEPT9 promoter were offset by losses in specificity, and therefore did not warrant development of final multiplex panel.
Development and clinical performance
The research studies above provided supportive data for a potential test based on the promoter methylation status of the SEPT9 in cell free DNA from plasma, using research methods for DNA extraction, bisulfite conversion and real time PCR [26]. A development project was undertaken to reduce the research observations to practice requiring standardization of each protocol step. The test was developed and optimized through different versions as outlined (Table 1) and the performance characteristics have been summarized in a meta-analysis including 25 studies [27].
| Test | Status | Sensitivity | Specificity | Comments |
|---|---|---|---|---|
| Epi proColon 1.0 | CE marked–no longer in use | 67% | 88% | Modified version used in PRESEPT study: 48% sensitivity, 91.5% specificity |
| Epi proColon 2.0 | CE marked, Chinese FDA approved | 73-81% | 96-99% | Primarily tested in Case Control Studies. Used in Europe and China. |
| Septin9 gene methylation assay | Chinese FDA approved | 77% | 96% | Tested in Opportunistic Screening setting, China |
| Epi proColon | US FDA PMA approved | 68-72% | 79-82% | Performance evaluation in Prospective Trial; compared to FIT in combined case control trial. USA |
Table 1: General test performance characteristics for the different versions of the Epi proColon test for the methylated SEPT9 promoter.
Epi proColon CE
The first test iteration was developed as Epi proColon CE for the European market. This test comprised magnetic particle based DNA isolation from 3.5 mLs plasma, bisulfite conversion using sodium bisulfite, repurification using magnetic particles, a duplex real-time PCR measuring actin beta as an internal DNA concentration control, and methylated SEPT9 promoter DNA as the target biomarker [28]. The PCR reaction is based on Heavy Methyl amplification using a blocker oligonucleotide to suppress amplification of non-methylated background DNA, combined with Methyl Light detection using a methylation specific detection probe [29]. Using this approach, the analytical sensitivity of the assay approached three genome equivalents per mL of plasma. The clinical performance was determined in a case control study of 261 patients, wherein 257 had valid tests (98.5%). The observed sensitivity for colorectal cancer was 69/103 (67%) patients with colorectal cancer, and importantly, early stage cancer detection was observed to be 44/66 (66.7%). The observed specificity from the no-evidence of disease class was 135/154 subjects (88%) who were negative for the test.
Prospective Evaluation of Septin 9 (PRESEPT) Study
PRESEPT was designed as a prospective clinical sample collection (ClinicalTrials.gov NCT00855348) to obtain plasma specimens from colorectal cancer screening subjects for whom a colonoscopy was performed as a reference method, and to evaluate the methylated SEPT9 promoter biomarker [30]. Each patient donated sufficient blood to isolate up to 20 mLs of plasma, which was bio-banked at -80°C in multiple aliquots. This prospective collection from 7941 individuals produced 6874 valid subjects, of which there were 53 colorectal cancer patients (CRC), 666 patients with advanced adenomas (AA), 2359 patients with non-advanced adenomas/polyps (NAA), and 3796 patients with no evidence of disease (NED). To evaluate the marker, all CRC and a stratified random sample of AA, NAA and NED patients were tested. Under the protocol, the testing comprised a duplicate PCR analysis. A post hoc analysis was performed in which a third PCR replicate was included for subjects with remaining available bisulfite converted DNA. This results in analysis of 45 μl of total bisulfite converted DNA, rather than 30 μl as used in the primary analysis. Sample identity was masked until completion of testing and data lock.
In the primary analysis, the observed sensitivity for colorectal cancer was 27/53 or 50.9%. This value was statistically adjusted to 48.2% to represent the patient distribution expected in the US population. The combined non-CRC detection rate was 126/1457 (8.6%) for a specificity of 91.4%, which was adjusted statistically to 91.5% to represent distribution expected in the US population. The post-hoc analysis was performed for 51 of the 53 CRCs and 1427 of the 1457 non CRCs based on DNA availability. In this analysis using a rule whereby a patient was positive if one of the three replicates was positive, the sensitivity was 63.9% at a specificity of 88.4%. This outcome indicates that the use of additional analyte increases test sensitivity at an acceptable cost in specificity [30].
Epi proColon 2.0 and the US FDA PMA approved Epi proColon
Based on experience with the original version of Epi proColon, a number of changes were made to the pre-analytic and analytic components and processes to further optimize the assay. These included: 1) Alternative magnetic particles to lower potential PCR inhibition; 2) The use of ammonium bisulfite rather than sodium bisulfite to allow inclusion of a liquid form of bisulfite as a kit component; 3) An increase in bisulfite conversion temperature and a shorter reaction time; 4) Modifications to the PCR oligonucleotides and fluorescent probes to optimize the reaction; 5) Three PCR replicates were performed per test to insure that the maximum available DNA was analyzed. Extensive analytical and technical verification was performed, and the assay was shown to detect at the level of a single genome equivalent per mL of plasma.
Epi proColon 2.0 CE: Technical aspects and clinical performance for version 2.0 of the test were recently summarized [31]. The optimized assay was first CE certified as Epi proColon 2.0 for the European and world market excluding the US. This CE version of the assay was validated using plasma samples collected with EDTA or CPDA collection tubes. The assay was validated on both the AB7500 PCR instrument (Thermo Fisher Scientific) and the LC480 instrument (Roche Applied Sciences). The CE version of the assay used an algorithm by which 2 of 3 PCR replicates were required to be positive in order to call a patient sample positive, emphasizing test specificity.
Test performance (Table 1) was evaluated in a number of case control studies in Europe and China [27,31-34]. As outlined in these studies, test sensitivity was reported to range from 73- 81% and test specificity was reported to range from 96-99%. A modified version of this test was developed in China, and tested in an opportunistic screening setting [34].
Epi proColon–US FDA PMA approved
Using the identical kit components and protocol described for Epi proColon 2.0 above, the optimized assay was submitted for regulatory approval in the US with an algorithm requiring only 1 of 3 PCR replicates to be positive to call a sample positive. The PMA process represents the highest regulatory standard for a diagnostic test, and required multiple clinical trials to provide sufficient evidence for approval.
Epi proColon Clinical Trials
Performance Evaluation: PMA approval requires extensive clinical validation in the intended use population. The study used the PRESEPT sample collection (ClinicalTrials.gov NCT00855348) [35] as outlined above, in which blood was prospectively collected from subjects at average risk for colorectal cancer who were referred for screening colonoscopy according to screening guidelines. The colonoscopy result was used as the reference standard. A stratified random sampling approach similar to the PRESEPT study was used, and methylated SEPT9 promoter was measured with the new Epi proColon test for all available CRC (44) and AA (621) patients, and a stratified random sample of 435 NAA, and 444 NED subjects. In this evaluation a raw sensitivity of 68% was observed at a specificity of 79%. When adjusted to population values the sensitivity remained 68% at a specificity of 80%. These data, using an equivalent evidence base, define the performance characteristics of the Epi proColon test, in comparison with the earlier version of the SEPT9 test evaluated in the PRESEPT study [30,35].
Comparison to fecal testing (ClinicalTrials.gov NCT01580540): As a requirement for approval, the US FDA also requested a study comparing performance to a commonly used stool blood test, the OC FIT-CHEK fecal immunochemical test. In this study, to compare sensitivity, stool and plasma samples were collected >10 days following colonoscopy, from cancer patients whose cancer was detected by a screening colonoscopy. This approach allowed the collection of a sufficiently large number of cases (100) that were representative of the prospective screening population, while limiting the collection bias. To compare specificity, stool and plasma samples were collected from prospective patients referred for screening colonoscopy, prior to bowel preparation [36]. In this study, the observed sensitivity and specificity for Epi proColon were 72 and 82% respectively compared with 68 and 97% for the OC FIT-CHEK test. On this basis, the sensitivity of the Epi proColon test was found to be non-inferior to the OC FITCHEK test. An interesting observation from this study was that FIT and Epi proColon testing were complimentary, with a combined sensitivity of 88.7% [36].
Adherence to Minimally Invasive Testing–ADMIT (ClinicalTrials. gov, ID NCT02251782): The FDA requested a final study to determine if patients would accept a blood based test. In the multicenter randomized ADMIT trial, acceptance of Epi proColon blood testing was 99.5%, compared with 88.1% for FIT, demonstrating a high degree of acceptance for the blood test [37]. In this study, 67% of patients with a positive test scheduled a follow-up diagnostic colonoscopy, and of those completing colonoscopy, 59% had a finding of a lesion (adenoma, polyp or other) requiring follow-up pathology.
Conclusion
Based on the clinical performance evaluation, the comparison with stool based FIT testing, and the observed degree of screening participation in the ADMIT trial, Epi proColon was approved by the US FDA as the first DNA methylation based test using cell free DNA in plasma as the analyte. Based on evidence from multiple trials [27] blood based screening for colorectal cancer is now available as a regulated product under CE labeling, and as an approved product under the Chinese FDA and the US FDA.
This DNA promoter methylation test, which has similar sensitivity to the stool based FIT test, addresses the clinical challenge of reaching patients who are unwilling or unable to be screened for colorectal cancer by other recommended methods. It is clear from numerous studies that despite screening recommendations, these other methods can present a significant barrier to screening [38]. Blood based testing using the SEPT9 promoter methylation test provides a viable option as reported in two independent trials. In a test choice trial in Germany, 83% of patients who refused a screening colonoscopy chose to be screened with the blood test, and 15% opted for a stool test, for a combined 98% coverage by non-invasive testing [39]. Furthermore in the randomized ADMIT trial in the US described above, 99.5% of patients who had previously not completed screening after at least two recommendations, proceeded to be screened with the blood test [37]. In this study, for the patients with a positive test, 67% scheduled a diagnostic colonoscopy within 3 months, and for those patients with a diagnostic colonoscopy, 59% had a finding resulting in a polypectomy or biopsy. These studies demonstrate the potential blood based testing has to close the screening gap.
Clearly, the test is not a replacement for colonoscopy, and patients with a positive test are referred for diagnostic colonoscopy. In the US, the test is indicated for patients who decline other approved screening methods. This addresses a very significant unmet need as one third of screening eligible people in the US; populations of ~23 million as estimated by the American Cancer Society, have not been screened or are not up to date according to screening guidelines. In this regard, this clinical epigenetic application has the potential to reach millions of people, help to identify colorectal cancer at an earlier treatable stage, and thereby save lives.