- (2011) Volume 12, Issue 4
Tamas A Gonda1*, Aimee Lucas1, Muhammad Wasif Saif2
1Division of Digestive and Liver Disease, 2Division of Hematology and Oncology;
Department of Medicine, Columbia University Medical Center. New York, NY, USA
Screening and early detection of pancreatic cancer has the potential to substantially impact outcomes in this deadly disease. Over the last ten years several cohort studies have been conducted and report on the yield of screening in high risk populations. With better understanding of the cellular compartments and the genetic and epigenetic changes that occur, biomarkers have also emerged as promising means of early detection. In this paper we summarize the results of the latest screening cohort and highlight a novel proteomic approach that may be used in future biomarker studies.
Mass Screening; Pancreatic carcinoma, familial; Pancreatic Neoplasms; Proteomics
EUS: endoscopic ultrasound; IPMN: intraductal papillary mucinous neoplasm; PanIN: pancreatic intraepithelial neoplasm; SILAC: stable isotope labeling by amino acids in cell culture
The majority of pancreatic cancer is discovered at late stage and is therefore incurable [1]. Recent recognition of precursor lesions and high risk populations suggests that screening for pancreatic cancer in highly selected populations may be appropriate [2]. Current recommendations suggest screening of high-risk individuals only under research protocols, with a cutoff of 10-fold, or more, increased risk of pancreatic cancer [3]. Over the last decade several groups have embarked on screening individuals with recognized genetic syndromes or a significant family history for pancreatic cancer [4]. Although there is substantial diversity among the screened populations these studies provide important information about the yield of these strategies. In addition, important genetic and epigenetic pathways have been recognized in the development of pancreatic cancer and these may provide better markers of cancer risk. These markers have mostly been found in whole tumor tissue, but recent advances clearly suggest that the different cell types that make up pancreatic cancer (epithelial cells, stromal fibroblasts, inflammatory cells, endothelial cells) may independently undergo genetic or epigenetic changes [5]. This suggests that cells of each tumor compartment may harbor different markers of carcinogenesis. In this summary we review results from a cohort of patients at risk for pancreatic cancer and present results of a novel screening approach focused on the tumor microenvironment.
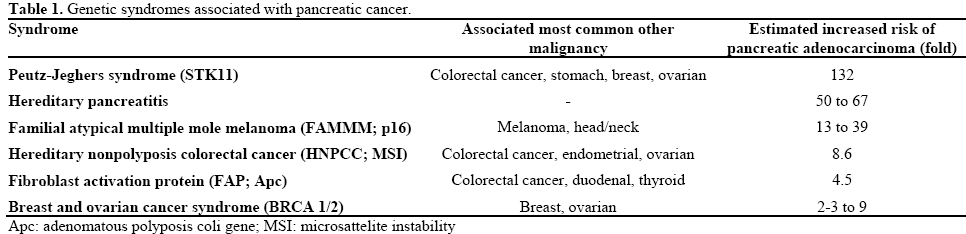
At this time, screening for pancreatic cancer is not recommended by society and national practice guidelines in the general population [3]. Individuals with a family history or with certain genetic syndromes have been considered targets of pancreatic cancer screening. Table 1 summarizes the common genetic syndromes that are associated with pancreatic cancer. In addition, a significant family history of pancreatic cancer seems to increase the risk of pancreatic cancer. Recent review of registries suggests that having one, two and three first degree relatives with pancreatic cancer translates into a 3.2, 6.4 and 32 fold increase in risk, respectively [7, 8]. The modalities used for screening are equally diverse and include cross sectional imaging (CT or MRI), endoscopic ultrasound (EUS) and testing for early glucose intolerance or serum markers (CA 19-9, CA 72) [9]. The longest follow up available is a ten year experience; however, the average length of time that one individual is surveyed in these programs is much shorter. Table 2 summarizes some of the results of recent screening programs in identifying significant pancreatic abnormalities.
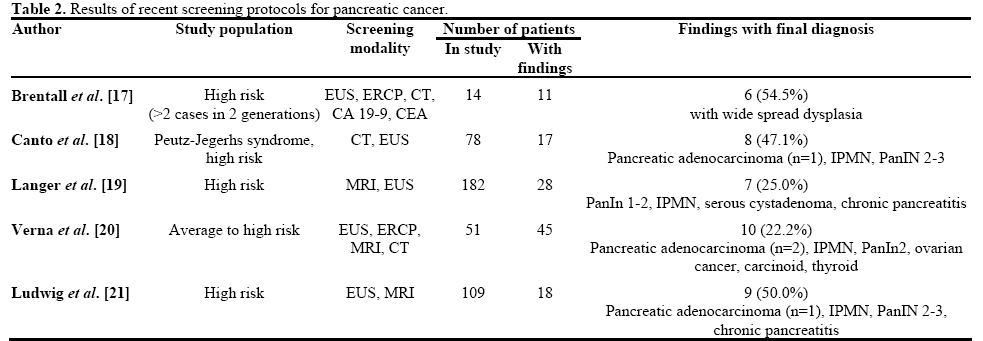
The study reported by Al-Sukhni et al. [6] adds to the growing number of registries of pancreatic screening program. They report on a population of 259 individuals judged to be at high risk of pancreatic cancer based n family history or a known genetic syndrome. The primary methods of screening used were MRI, which prompted further investigation with EUS or biopsy only if an abnormality was found. It is important to note that this is a slightly different strategy from other screening programs where EUS often complements cross sectional imaging. MRI was performed annually in this protocol. The average length of follow up was 3.2 years, although the majority of findings were found at the initial evaluation or at the first interval evaluation. Seventeen abnormalities were noted, the majority of which were branch duct IPMNs (n=15). However, 2 out of 259 patients developed advanced pancreatic cancer during the study period.
The overall yield of this study is similar to several of the prior registry data and suggests that IPMNs are the most common pancreatic abnormalities recognized in these patients. Further and longer term studies will need to evaluate whether IPMN in these patients represents a higher than usual risk of progression. As outlined in Table 2 the diagnosis of an occult cancer is rare in this population. Unfortunately both patients in this study died of their disease and this raises an important question whether early detection can be translated into a survival benefit.
The tumor microenvironment that consists of the inflammatory cells, endothelial cells and the cancer associated fibroblasts, have increasingly been recognized to play an active role in carcinogenesis. Cancer associated fibroblasts have been shown to have distinct genetic and epigenetic characteristics and are therefore emerging as therapeutic targets and sources of biomarkers [5, 11]. In pancreatic cancer, one of the most stroma abundant cancer types, cancer associated fibroblasts may be in part derived from the pancreatic stellate cells. Activation of pancreatic stellate cells is seen in both cancer and pancreatitis and it is thought that paracrine signaling from these cells and therefore secreted proteins may impact cancer progression. The interplay between epithelial cells and pancreatic stellate cells [12] is likely important to maintain both phenotypes as shown in Figure 1.
Figure 1. Factors responsible for the activation of pancreatic stellate cells and the interaction between epithelial cells and stromal cells. The activation of quiescent pancreatic stellate cells (qPSCs, which are characterized by the expression of glial fibrillary astrocytic protein (GFAP) and desmin) into activated stellate cells (aPSC which are characterized by alpha-smooth muscle actin (ASMA) expression) is stimulated by multiple chemokines such as interleukins 1 and 6 (IL1, 6) and tumor necrosis factor-alpha (TNFα) and tumor cell derived factors such as platelet-derived growth factor (PDGF), transforming growth factor-beta (TGFβ) and fibroblast growth factor-2 (FGF2). Activated pancreatic stellate cells (aPSC) also contribute to the tumor epithelial cell phenotype.
Earlier work by Wehr et al. [13] has characterized the proteome that distinguishes quiescent and activated stellate cells and validated several of these differentially expressed proteins in human pancreatic cancer sections by immunohistochemistry. In their current study they use this similar method and label these secreted proteins using a cell based protein labeling system (stable isotope labeling by amino acids in cell culture, SILAC). This technique was originally developed to allow the simultaneous detection of many secreted proteins by different cell populations using radiolabeling with 12C- and 13C-labeled amino acids [14]. After incorporation of one or the other isotope and mixing of the lysates, tandem mass spectrometry is used to identify differential proteins [15]. In their study SILAC labeling was used to identify activated pancreatic stellate cell derived proteins and these proteins were further interrogated for their potential as serum biomarkers using multiple-reaction monitoring mass spectrometry (MRM-MS) [16].
The study identified 40 pancreatic stellate cell secreted proteins in serum. One-hundred and eighty-three unique peptides corresponded to these proteins and of these 69 peptides (corresponding to 36 proteins could be validated by mass spectrometry. Their findings were validated on 10 human pancreatic cancer cases and controls. The results of this presentation are preliminary and suggest that this method may be a promising way to identify novel biomarkers using a proteomic approach.
These studies highlight two important areas of investigation in pancreatic cancer screening and early detection. Data from somewhat diverse screening programs suggest a low to moderate yield in identifying at risk individuals. However, the period of observation is limited in these studies and therefore it is difficult to judge the clinical significance of findings. The importance of better biomarkers is emerging as a critical area for effective pancreatic cancer screening. Proteomic approaches maybe one of the most valuable tools to identify serum and stool based markers and the combined strategy described above allows for a powerful tool to accomplish this goal.
None