Keywords
Chaperones; Co-chaperones; Heat-shock response; HSF1; Hsp90; Posttranslational regulation
Introduction
Hsp90α and Hsp90β interact with ~2000 client proteins [1,2], although only ~725 of these have been confirmed by direct protein-protein interaction. Hsp90 consists of an N-terminal ATP binding domain, separated from the middle domain by a charged linker (Figure 1). The C-terminal domain is not only responsible for the inherent dimerization of the chaperone, but the conserved MEEVD motif, at its extreme C-terminus, provides the binding site for a variety of TPR domain containing co-chaperones. These include the phosphatase Pp5, immunophilins (for example FKBP51 and FKBP52) and HOP, that aid and modulate its function.
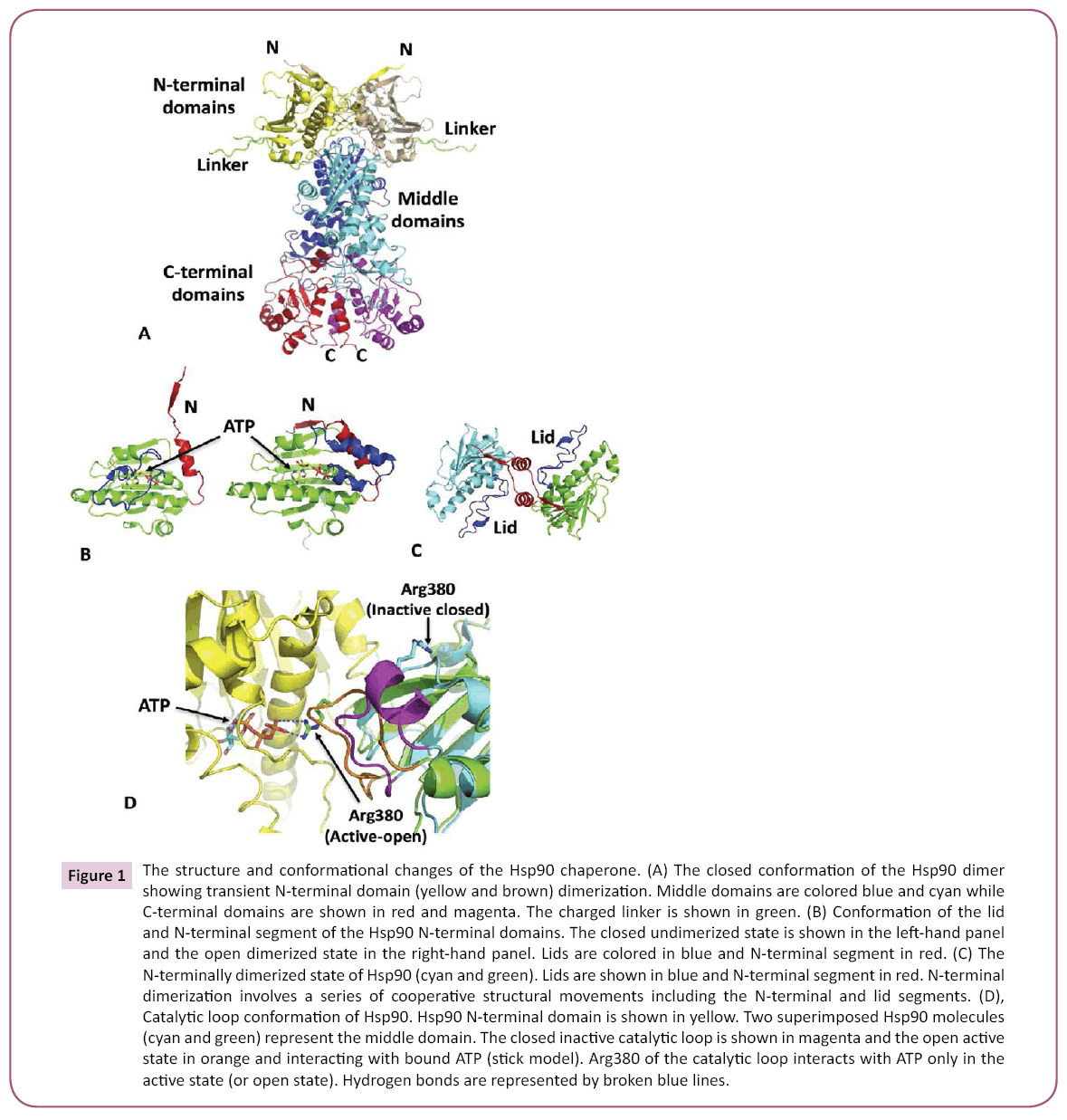
Figure 1: The structure and conformational changes of the Hsp90 chaperone. (A) The closed conformation of the Hsp90 dimer showing transient N-terminal domain (yellow and brown) dimerization. Middle domains are colored blue and cyan while C-terminal domains are shown in red and magenta. The charged linker is shown in green. (B) Conformation of the lid and N-terminal segment of the Hsp90 N-terminal domains. The closed undimerized state is shown in the left-hand panel and the open dimerized state in the right-hand panel. Lids are colored in blue and N-terminal segment in red. (C) The N-terminally dimerized state of Hsp90 (cyan and green). Lids are shown in blue and N-terminal segment in red. N-terminal dimerization involves a series of cooperative structural movements including the N-terminal and lid segments. (D), Catalytic loop conformation of Hsp90. Hsp90 N-terminal domain is shown in yellow. Two superimposed Hsp90 molecules (cyan and green) represent the middle domain. The closed inactive catalytic loop is shown in magenta and the open active state in orange and interacting with bound ATP (stick model). Arg380 of the catalytic loop interacts with ATP only in the active state (or open state). Hydrogen bonds are represented by broken blue lines.
Binding of ATP results in a coordinated restructuring of Hsp90 [3], including the release of an N-terminal domain lid-region, which binds over the N-terminal domain bound ATP and simultaneously provides a platform for N-terminal dimerization (Figure 1). Additionally, the middle-domain catalytic loop of Hsp90 is able to adopt an active conformation by interacting with bound ATP (Figure 1). These structural changes together conspire in a coordinated fashion to form an N-terminally dimerized state that is catalytically active [4-6] (Figure 1). The dimerized state is further stabilized by a β-strand swap involving the N-terminal domains (Figure 1). A detailed description of these changes has been previously described [3,7]. Importantly, the varied structural changes of Hsp90 allow for a variety of regulatory points that can affect the Hsp90 chaperone cycle. These include both PTMs and modulation by co-chaperones. However, the rate-limiting step of the chaperone cycle is determined by the structural rearrangements of Hsp90, which ultimately ends with the hydrolysis of ATP and the disassociation of N-terminal dimerization, and perhaps maturation of a client protein.
Transcriptional Regulation and Heat Shock Response
HSF1, which is itself a Hsp90 dependent client protein, is the master regulator of the heat shock response (HSR) [8-11] and consequently under strict regulation. Determining the mechanistic action of HSF1 in detail is central to understanding the complex transcriptional regulatory systems of Hsp90. HSF1 is normally associated with Hsp90 in a repressed state, but during stress HSF1 is released [12-14] and homotrimerises [14], whereupon it gains DNA binding activity and is targeted to heatshock- factor elements (HSE). However, to gain transcriptional activity and transactivation competence [15-23], a series of phosphorylations are required (notably at Ser 230) [24,25], which leads to a rapid up-regulation of Hsp90, Hsp70 and a variety of co-chaperones including Hsp27 and Hsp40 [26]. Additionally, stress is required to overcome a stress-regulated repressive state brought about by specific phosphorylations (Ser 230, Ser 303 and Ser 307) [24,27,28], and HSF1 is known to respond directly to changes in temperature [29-31]. Phosphorylations of HSF1 are also known to be involved in the integration of signals from other signaling pathways [24,25]. For example, phosphorylation at Ser 326 promotes association with Daxx, a mediator of HSF1 activation. Although the phosphorylation of HSF1 is important it appears that the stress-inducible phosphorylation of HSF1 can be uncoupled from its activation [32].
SUMOylation [33-36] and acetylation [37-39] are reported to regulate HSF1. A number of other mechanisms also appear to regulate HSF1, including heat stress, which is directly detected by the regulatory domain of HSF1 [40], by Hsp90 repression of trimeric HSF1 [12,41], and by the inhibition of HSF1 transactivating activity by Hsp70 and Hsp40. The inhibitory activity of Hsp70 is probably through the recruitment of CoREST (corepressor for element-1-silencing transcription factor), a transcriptional co-repressor of Hsp70 [42,43]. An activator consisting of the eukaryotic elongation factor 1A (eEF1A) together with a constitutively expressed non-coding heat-shock RNA-1 RNA has been reported to upregulate HSF1 [44,45]. The molecular chaperone TriC has also been seen to associate with HSF1 [46], however, the significance of this remains unknown.
Active HSF1 associates with HSE that consist of a number of nGAAn units (reviewed in [47]) where their exact spatial arrangement can influence cooperativity between binding trimers of HSF. The promotor regions of Hsp90 genes are complex providing elements that downregulate and upregulate expression [48] (Figure 2), STAT-1 and STAT-3 binding sites appear to overlap with HSE of HSF1 [47,49,50]. Strong transcriptional activation is seen by the interaction between HSF1 and STAT-1. In contrast, HSF1 and STAT-3 antagonize each other because they appear unable to interact with each other, and thus reduce expression of Hsp90β [49,51]. IL (interleukin)-6 transcription factor NF-IL6 (nuclear factor for IL-6) can also upregulate Hsp90β. On the other hand, Hsp90 β regulation is augmented by the stress-responsive activator of p300, Strap [52], which together form a HSF1 chromatin-associated complex. Since p300 has been reported to possess a histone acetylase activity [53], chromatin acetylation may be a mechanism by which Hsp90 expression is upregulated. The 5′-flanking region of the HSP90AA1 promoter (but not in HSP90AB1) is also bound by NF-B [54], and the dependence of NF-B and IKK (inhibitor of NF-B kinase) on Hsp90 suggests a regulatory loop that can influence a cells response to stress and ultimately its survival.
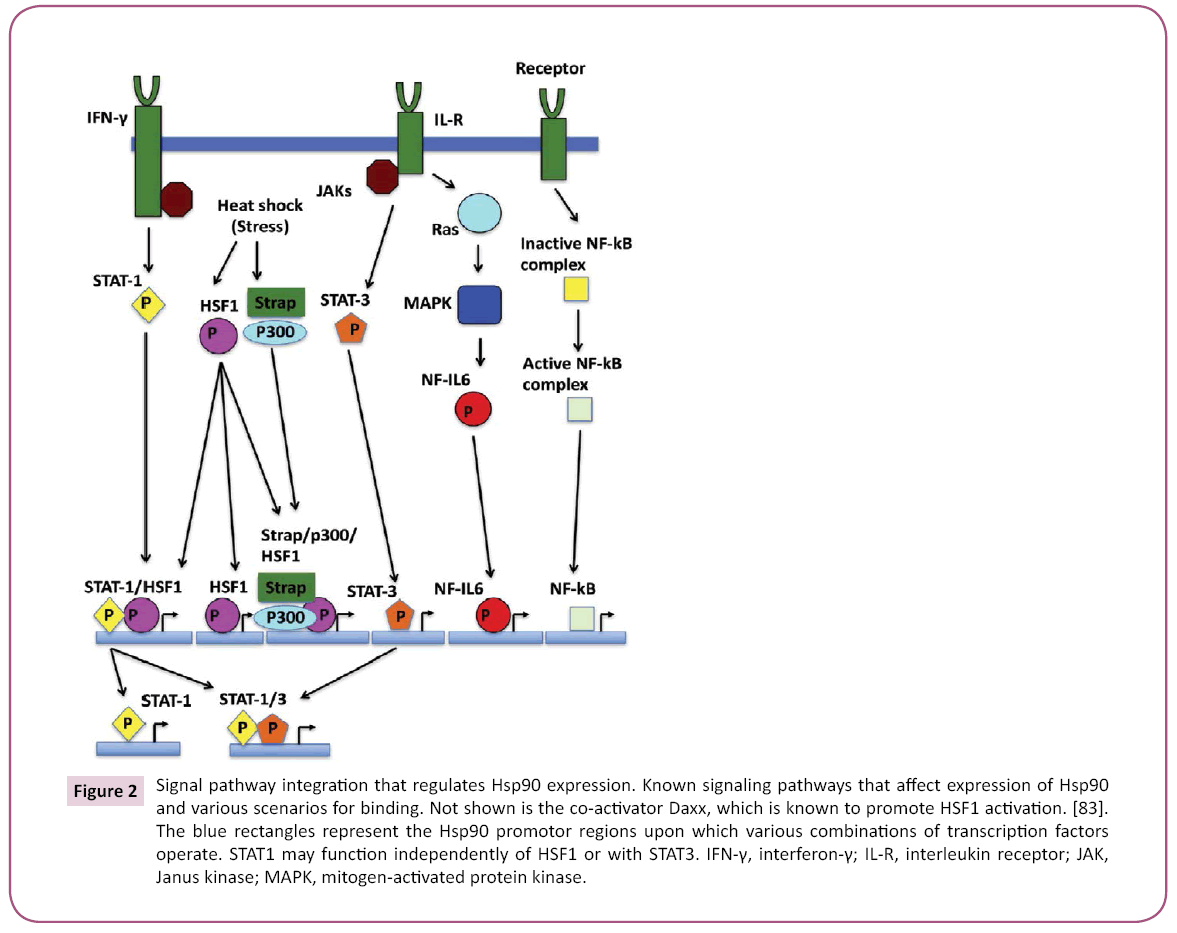
Figure 2: Signal pathway integration that regulates Hsp90 expression. Known signaling pathways that affect expression of Hsp90 and various scenarios for binding. Not shown is the co-activator Daxx, which is known to promote HSF1 activation. [83]. The blue rectangles represent the Hsp90 promotor regions upon which various combinations of transcription factors operate. STAT1 may function independently of HSF1 or with STAT3. IFN-γ, interferon-γ; IL-R, interleukin receptor; JAK, Janus kinase; MAPK, mitogen-activated protein kinase.
Clearly, the regulation of transcription of Hsp90 represents a central hub at which diverse signals can be integrated into regulating Hsp90 levels and the HSR (Figure 2). In order to integrate such diverse signals, HSF1 plays a major role and is consequently itself subject to a complex regulatory process, including the ability to to directly sense heat stress.
Post-Translational Regulation of The Hsp90 Complex
PTM of Hsp90 and its co-chaperones include not only phosphorylation, but also acetylation, methylation, S-nitrosylation, SUMOylation and ubiquitylation and have been reviewed in [55,56]. Such modifications have been shown to be specific to either Hsp90α or Hsp90β [57,58], and can regulate Hsp90 activity either directly or by its interaction with co-chaperones, nucleotides or client protein [57,59-63]. PTM of co-chaperones has been shown to be necessary for the chaperoning of kinase clients [64-66] and Ser 13 dephosphorylation of Cdc37p50 by PP5/Ppt1 appears to signal chaperone cycle progression [67]. In contrast, Cdc37p50 phosphorylation at Tyr 4 and Tyr 298 appears to disrupt Cdc37p50-client association and thus provides directionality to the chaperone cycle [61]. Additionally, Tyr197 phosphorylation of Hsp90 appears to cause Cdc37p50 dissociation from Hsp90 [61], whereas Tyr 313 phosphorylation may promote the recruitment of Aha1, both of which stimulate the ATPase activity of Hsp90 and further the chaperoning process. c-Abl kinase has been reported to phosphorylate of Tyr 223 of human Aha1, which appears to differentially affect client protein association [68], however, the same authors reported that Tyr 223 phosphorylation also led to proteasome degradation of Aha1. Tyr 627 phosphorylation of Hsp90α induces client and co-chaperones dissociation, which might signal completion of the kinase chaperone cycle.
The dimerization of Sgt1, another Hsp90 co-chaperone, appears to be influenced by Ser 361 phosphorylation. This in turn affects kinetochore assembly and therefore chromosome segregation in eukaryotic cell division [69]. p23 (cytoplasmic prostaglandin E synthase 3) [70], murine Sti1/HOP and FKBP52 are other Hsp90 co-chaperones that have been shown to be regulated by PTMs, and have roles in a variety of processes including the cell cycle, steroid hormone activation and telomerase maturation [65,71-74].
Clearly, PTM of Hsp90 and its co-chaperones are a major regulatory mechanism of the chaperone cycle, such that the activation of specific client proteins is optimized. This is critically important as the clientele of Hsp90 collectively represent a structurally diverse set of proteins, whose maturation and activation have their own specific requirements.
Regulation of Hsp90 by Co-Chaperones
The chaperone cycle of Hsp90 is driven by coordinated structural rearrangements following ATP binding, which leads to N-terminal dimerization of Hsp90 [3,5,7,75]. The Co-chaperones HOP, Sgt1 and Cdc37p50 are major players in delivering client proteins to the Hsp90 chaperone (see [7,76] for reviews). Cdc37p50 and HOP silence Hsp90 ATPase activity and thus facilitate the binding of client protein with Hsp90 [77-79]. In contrast, Sgt1 when in complex with the plant protein Rar1 (CHORD domain containing co-chaperone, also known as Chp1 and melusin in mammals) may form a stable ADP bound complex [80].
Other co-chaperones help to regulate the chaperone cycle by influencing the ATPase activity of Hsp90. For example, Aha1 strongly accelerates the ATPase activity of Hsp90 [6,81] by promoting the rate-limiting structural changes of Hsp90 [3,75]. In contrast, the ATPase activity of Hsp90 is slowed by the cochaperone Sba1 [81], although the human orthologue p23 shows a more robust inhibition [82]. The slowing of the cycle may be important for generating longer lived client-Hsp90 complexes, which may favor their maturation.
In summary, co-chaperone modulation of the chaperone cycle is clearly an important mechanism by which specific Hsp90 complexes are regulated, and the complex set of structural changes of Hsp90 has allowed a variety of mechanisms to evolve that act upon these critically important points of structural change (Figure 3).
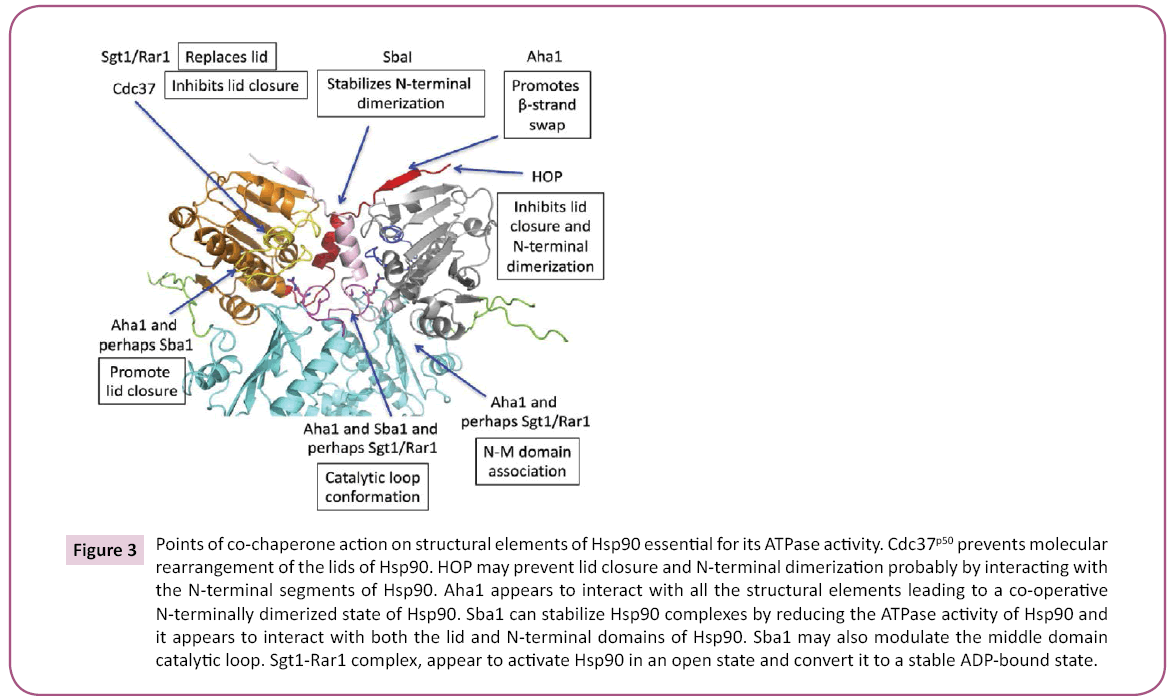
Figure 3: Points of co-chaperone action on structural elements of Hsp90 essential for its ATPase activity. Cdc37p50 prevents molecular rearrangement of the lids of Hsp90. HOP may prevent lid closure and N-terminal dimerization probably by interacting with the N-terminal segments of Hsp90. Aha1 appears to interact with all the structural elements leading to a co-operative N-terminally dimerized state of Hsp90. Sba1 can stabilize Hsp90 complexes by reducing the ATPase activity of Hsp90 and it appears to interact with both the lid and N-terminal domains of Hsp90. Sba1 may also modulate the middle domain catalytic loop. Sgt1-Rar1 complex, appear to activate Hsp90 in an open state and convert it to a stable ADP-bound state.
Conclusion
Many biological processes, including stress adaptation are dependent on Hsp90 and numerous regulatory systems therefore operate to integrate and regulate Hsp90 activity appropriately. As a master regulator of the HSR, HSF1 not only helps to determine Hsp90 levels directly but integrates varied cellular signals into the transcriptional control Hsp90. Direct regulation of the Hsp90 protein involves PTMs, as well as an ability to directly sense heat stress and by direct control through co-chaperone action and its client proteins, which are themselves subject to various regulatory processes. Although some progress has been made in understanding these processes, a substantial amount remains unknown and our knowledge of Hsp90 regulation remains in its infancy. Many of the regulatory enzymes, including phosphatases, kinases, histone deacetylases and histone acetylases remain unknown. Determining how such modifications are translated into coherent regulatory processes will be challenging, but is non-the-less essential to understanding the Hsp90 chaperone cycle and the various biological processes dependent on Hsp90.
Acknowledgment
This work was funded by the Wellcome Trust [grant number 095605/Z11/Z]. I thank their financial support.
References
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, et al. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715-727.
- Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D (2011) An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One 6: e26044.
- Schulze, A, Beliu G, Helmerich DA, Schubert J, Pearl LH, et al.(2016) Cooperation of local motions in the Hsp90 molecular chaperone ATPase mechanism. Nat ChemBiol 12: 628-635.
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, et al. (2000) The ATPase cycle of Hsp90 drives a molecular 'clamp' via transient dimerization of the N-terminal domains. EMBO Journal 19: 4383-4392.
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, et al.(2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440: 1013-1017.
- Meyer P, Prodromou C, Liao C, Hu B, Roe SM, et al.(2004) Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J 23: 511-519.
- Prodromou C(2012) The 'active life' of Hsp90 complexes. BiochimBiophysActa 1823: 614-623.
- Voellmy R(2004) On Mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress & Chaperones 9: 122-133.
- Sorger PK, Pelham HR (1988) Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54: 855-864.
- Pirkkala L, Nykanen P, Sistonen L (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118-1131.
- Wiederrecht G, Seto D, Parker CS (1988) Isolation of the gene encoding the S. cerevisiae heat shock transcription factor. Cell 54: 841-853.
- Ali A, Bharadwaj S, O'Carroll R, Ovsenek N(1988) HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes.Mol Cell Biol 18: 4949-4960.
- Bharadwaj S, Ali A, Ovsenek N (1999) Multiple components of the HSP90 chaperone complex function in regulation of heat shock factor 1 in vivo. Mol Cell Biol 19: 8033-8041.
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R (1998) Repression of heat shock transcription factor HSF1 activation by Hsp90 (Hsp90 complex) that forms a stress-sensitive complex with HSF1. Cell 94: 471-480.
- Amin J, Ananthan J, Voellmy R (1988) Key features of heat shock regulatory elements. Mol Cell Biol 8: 3761-3769.
- Pelham HR (1982) A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30: 517-528.
- Xiao H, Lis JT (1988) Germline transformation used to define key features of heat-shock response elements. Science 239: 1139-1142.
- Cotto JJ, Kline M, Morimoto RI (1996) Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J BiolChem 271: 3355-3358.
- Jurivich DA, Sistonen L, Kroes RA, Morimoto RI (1992) Effect of sodium salicylate on the human heat shock response. Science 255: 1243-1245.
- Zuo J, Rungger D, Voellmy R (1995) Multiple layers of regulation of human heat shock transcription factor 1.Mol Cell Biol 15: 4319-4330.
- Bruce JL, Price BD, Coleman CN, Calderwood SK(1993) Oxidative injury rapidly activates the heat shock transcription factor but fails to increase levels of heat shock proteins. Cancer Res 53: 12-15.
- Hensold JO, Hunt CR, Calderwood SK, Housman DE, Kingston RE(1990) DNA binding of heat shock factor to the heat shock element is insufficient for transcriptional activation in murine erythroleukemia cells.Mol Cell Biol 10: 1600-1608.
- McConnell JR, Buckton LK, McAlpine SR (2015) Regulating the master regulator: Controlling heat shock factor 1 as a chemotherapy approach. BioorgMed ChemLett 25: 3409-3414.
- Anckar J, Sistonen L (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease.Annu Rev Biochem 80: 1089-1115.
- Holmberg CI, Tran SE, Eriksson JE, Sistonen L(2002) Multisite phosphorylation provides sophisticated regulation of transcription factors. Trends BiochemSci 27: 619-627.
- Heimberger T, Andrulis M, Riedel S, Stühmer T, Schraud H, et al. (2013) The heat shock transcription factor 1 as a potential new therapeutic target in multiple myeloma. Br J Haematol 160: 465-476.
- Kline MP, Morimoto RI (1997) Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation.Mol Cell Biol 17: 2107-2115.
- Knauf U, Newton EM, Kyriakis J, Kingston RE(1996) Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev 10: 2782-2793.
- Bulman AL, Nelson HC (2005) Role of trehalose and heat in the structure of the C-terminal activation domain of the heat shock transcription factor. Proteins 58: 826-835.
- Pattaramanon N, Sangha N, Gafni A (2007) The carboxy-terminal domain of heat-shock factor 1 is largely unfolded but can be induced to collapse into a compact, partially structured state. Biochemistry 46: 3405-3415.
- Sorger PK (1990) Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62: 793-805.
- Budzyński MA, Puustinen MC, Joutsen J, Sistonen L(2015) Uncoupling Stress-Inducible Phosphorylation of Heat Shock Factor 1 from Its Activation. Mol Cell Biol 35: 2530-2540.
- Anckar J, Sistonen L (2007) SUMO: Getting it on. BiochemSoc Trans 35: 1409-1413.
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, et al.(2006) PDSM, a motif for phosphorylation-dependent SUMO modification. ProcNatlAcadSci USA 103: 45-50.
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947-956.
- Newton EM, Knauf U, Green M, Kingston RE(1996) The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress.Mol Cell Biol 16: 839-846.
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI (2009) Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323: 1063-1066.
- Liu PC, Thiele DJ (1999) Modulation of human heat shock factor trimerization by the linker domain. J BiolChem 274: 17219-17225.
- Vujanac M, Fenaroli A, Zimarino V (2005) Constitutive nuclear import and stress-regulated nucleocytoplasmic shuttling of mammalian heat-shock factor 1. Traffic 6: 214-229.
- Brown SA, Weirich CS, Newton EM, Kingston RE (1998) Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J, 1998. 17(11): p. 3146-54.
- Morimoto RI (2002) Dynamicremodeling of transcription complexes by molecular chaperones. Cell 110: 281-284.
- Shi Y, Mosser DD, Morimoto RI (1998) Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12: 654-666.
- Gómez AV, Galleguillos D, Maass JC, Battaglioli E, Kukuljan M, et al.(2008) CoREST represses the heat shock response mediated by HSF1. Mol Cell 31: 222-231.
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E (2006) RNA-mediated response to heat shock in mammalian cells. Nature 440: 556-560.
- Kugel JF, Goodrich JA (2006) Beating the heat: A translation factor and an RNA mobilize the heat shock transcription factor HSF1. Mol Cell 22: 153-154.
- Neef DW, Turski ML, Thiele DJ (2010) Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoSBiol 8: e1000291.
- Sakurai H, Enoki Y (2010) Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J 277: 4140-4149.
- Prodromou C (2016) Mechanisms of Hsp90 regulation. Biochem J 473: 2439-2452.
- Stephanou A, Latchman DS (1999) Transcriptional regulation of the heat shock protein genes by STAT family transcription factors. Gene Expr 7: 311-319.
- Ehret GB, Reichenbach P, Schindler U, Horvath CM, Fritz S, et al.(2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J BiolChem276: 6675-6688.
- Stephanou A, Isenberg DA, Nakajima K, Latchman DS(1999) Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90beta gene promoters. J BiolChem 274: 1723-1728.
- Xu D, Zalmas LP, La Thangue NB (2008) A transcription cofactor required for the heat-shock response. EMBO Rep 9: 662-669.
- Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, et al. (2001) p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 20: 1331-1340.
- Ammirante M, Rosati A, Gentilella A, Festa M, Petrella A, et al.(2008) The activity of hsp90 alpha promoter is regulated by NF-kappa B transcription factors. Oncogene, 2008. 27: 1175-1178.
- Zuehlke AD, Beebe K, Neckers L, Prince T(2015) Regulation and function of the human HSP90AA1 gene. Gene 570: 8-16.
- Mollapour M, Neckers L (2012) Post-translational modifications of Hsp90 and their contributions to chaperone regulation. BiochimBiophysActa 1823: 648-655.
- Quanz M, Herbette A, Sayarath M, DeKoning L, Dubois T, et al. (2012) Heat shock protein 90alpha (Hsp90alpha) is phosphorylated in response to DNA damage and accumulates in repair foci. J BiolChem287: 8803-8815.
- Solier S, Kohn KW, Scroggins B, Xu W, Trepel J, et al. (2012) Heat shock protein 90alpha (HSP90alpha), a substrate and chaperone of DNA-PK necessary for the apoptotic response. ProcNatlAcadSci USA 109: 12866-12872.
- Mollapour M, Tsutsumi S, Neckers L (2010) Hsp90 phosphorylation, Wee1 and the cell cycle. Cell Cycle, 2010. 9: 2310-2316.
- Zhao YG, Gilmore R, Leone G, Coffey MC, et al.(2001) Hsp90 phosphorylation is linked to its chaperoning function. Assembly of the reovirus cell attachment protein. J BiolChem 276: 32822-32827.
- Xu W, Mollapour M, Prodromou C, Wang S, Scroggins BT, et al.(2012) Dynamic tyrosine phosphorylation modulates cycling of the HSP90-P50(CDC37)-AHA1 chaperone machine. Mol Cell 47: 434-443.
- Muller P, Ruckova E, Halada P, Coates PJ, Hrstka R, et al.(2013) C-terminal phosphorylation of Hsp70 and Hsp90 regulates alternate binding to co-chaperones CHIP and HOP to determine cellular protein folding/degradation balances. Oncogene 32: 3101-3110.
- Mollapour M, Tsutsumi S, Truman AW, Xu W, Vaughan CK,et al.(2011) Threonine 22 phosphorylation attenuates Hsp90 interaction with cochaperones and affects its chaperone activity. Mol Cell 41: 672-681.
- Shao J, Prince T, Hartson SD, Matts RL(2003) Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J BiolChem 278: 38117-38120.
- Miyata Y (2009) Protein kinase CK2 in health and disease: CK2: the kinase controlling the Hsp90 chaperone machinery. Cell Mol Life Sci 66: 1840-1849.
- Bandhakavi S, McCann RO, Hanna DE, Glover CV(2003) A positive feedback loop between protein kinase CKII and Cdc37 promotes the activity of multiple protein kinases. J BiolChem 278: 2829-2836.
- Vaughan CK, Mollapour M, Smith JR, Truman A, Hu B, et al.(2008) Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell 31: 886-895.
- Dunn DM, Woodford MR, Truman AW, Jensen SM, Schulman J, et al.(2015) c-Abl Mediated Tyrosine Phosphorylation of Aha1 Activates Its Co-chaperone Function in Cancer Cells. Cell Rep 12: 1006-1018.
- Bansal PK, Mishra A, High AA, Abdulle R, Kitagawa K(2009) Sgt1 dimerization is negatively regulated by protein kinase CK2-mediated phosphorylation at Ser361. J BiolChem, 2009. 284: 18692-18698.
- Kobayashi T, Nakatani Y, Tanioka T, Tsujimoto M, Nakajo S, et al.(2004) Regulation of cytosolic prostaglandin E synthase by phosphorylation.Biochem J 381: 59-69.
- Johnson JL, Toft DO (1994) A novel chaperone complex for steroid receptors involving heat shock proteins, immunophilins, and p23. J BiolChem 269: 24989-24993.
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, et al.(1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes & Development 13: 817-826.
- Longshaw VM, Dirr HW, Blatch GL, Lässle M (2000) The in vitro phosphorylation of the co-chaperone mSTI1 by cell cycle kinases substantiates a predicted casein kinase II-p34cdc2-NLS (CcN) motif. BiolChem 381: 1133-1138.
- Miyata Y, Chambraud B, Radanyi C, Leclerc J, Lebeau MC, et al.(1997) Phosphorylation of the immunosuppressant FK506-binding protein FKBP52 by casein kinase II: regulation of HSP90-binding activity of FKBP52. ProcNatlAcadSci USA 94: 14500-14505.
- Siligardi G, Hu B, Panaretou B, Piper PW, Pearl LH, et al.(2004) Co-chaperone regulation of conformational switching in the Hsp90 ATPase cycle. J BiolChem 279: 51989-51998.
- Prodromou C, Morgan RML (2016) "Tuning" the ATPase activity of Hsp90. AdvBiochem Health D 14: 469-490.
- Prodromou C, Siligardi G, O'Brien R, Woolfson DN, Regan L, et al.(1999) Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. Embo J 18: 754-762.
- Li J, Richter K, Buchner J (2011) Mixed Hsp90-cochaperone complexes are important for the progression of the reaction cycle. Nat StructMolBiol 18: 61-66.
- Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, et al. (2002) Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J BiolChem 277: 20151-20159.
- Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH(2010) Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: implications for chaperoning of NLR innate immunity receptors. Mol Cell 39: 269-281.
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, et al.(2002) Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell 10: 1307-1318.
- McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, et al.(2006) The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J MolBiol 356: 746-58.
- Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, et al. (2004) DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. ProcNatlAcadSci USA 101: 4100-4105.




