Review Article - (2016) Volume 2, Issue 2
Koichi Suzuki* and Toshiki Rikiyama
Department of Surgery, Saitama Medical Center, Jichi Medical University, 1-847 Amanuma-cho, Omiya-ku, Saitama 330- 8503, Japan
*Corresponding Author:
Koichi Suzuki
Department of Surgery, Saitama Medical Center
Jichi Medical University, 1-847 Amanuma-cho
Omiya-ku, Saitama 330- 8503, Japan.
Tel: +81-48-647-2111
Fax: +81-48-644-8617
Email: ksuzbnhm@yahoo.co.jp
Received date: April 22, 2016; Accepted date: April 30, 2016; Published date: May 06, 2016
Citation: Suzuki K, Rikiyama T. Pitfalls of KRAS Testing for Treating Patie ts with Metastatic Colorectal Cancer. J Clin Epigenet. 2016, 2:2. DOI: 10.21767/2472-1158.100018
Copyright: © 2016 Suzuki K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Background: KRAS testing is a critical initial step of the treatment for metastatic colorectal cancer patients. However, recent clinical trials have raised the issue of whether patient selection as determined by KRAS testing is appropriate.
Method: Here, we reviewed guidelines, literatures and clinical trials to elucidate whether detection threshold of KRAS testing is appropriate.
Results: It has been recommended that a detection threshold within 1–10% should be considered for RAS testing in Japan, but there is insufficient evidence for widespread adoption in the guideline. In literatures, retrospective studies have shown that high-sensitivity KRAS testing is more effective in the secondor third-line setting but no study has been conducted in the first-line setting. High-sensitivity KRAS testing in recent clinical trials has revealed undetectable mutant clones in tumors before chemotherapy. These clones have been shown to influence the outcome of second-line treatment, but there is no evidence of their influence in the first-line setting. On the other hands, there are accumulating data showing an additional effect of anti-epidermal growth factor receptor (EGFR) antibodies in patients harboring minor KRAS mutations determined by a low detection threshold of 5–10% in the first-line setting. These findings indicate that high-sensitivity KRAS testing have enabled to select super-responders, but may lead to exclusion of patients who could benefit from drug therapy.
Conclusion: In order not to deprive patients of the chance to benefit from anti- EGFR antibodies, the optimal cut-off value for KRAS testing should be determined for selection of patients likely to benefit from treatment in the first-line setting.
Introduction
Recent clinical trials have raised the issue of whether patient selection as determined by biomarker detection is appropriate. This selection has been shown to improve treatment outcomes and emphasizes the power of the drug advertising, however, selection also has the risk of excluding patients who might benefit from drug therapy (Figure 1A).
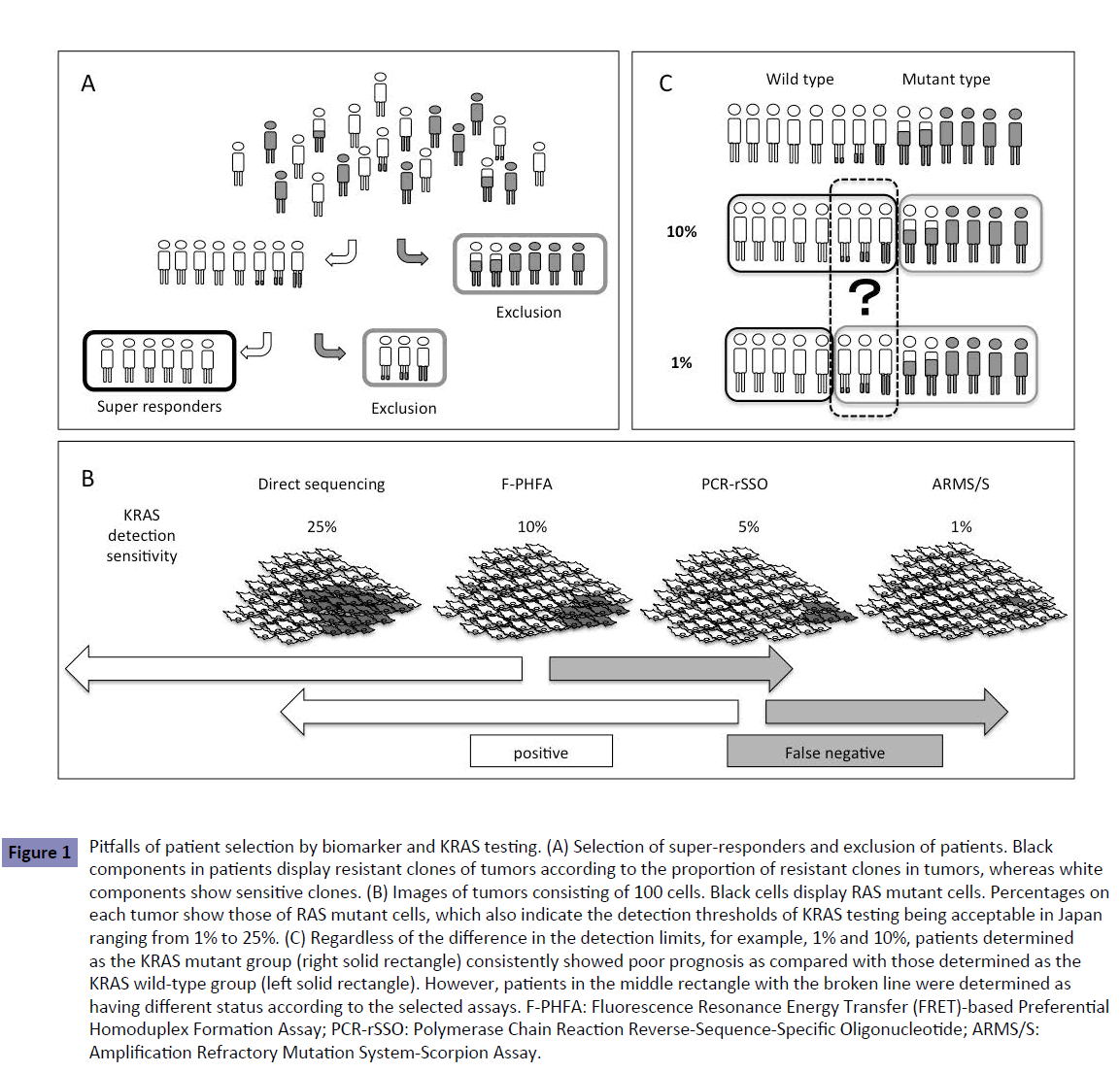
Figure 1: Pitfalls of patient selection by biomarker and KRAS testing. (A) Selection of super-responders and exclusion of patients. Black components in patients display resistant clones of tumors according to the proportion of resistant clones in tumors, whereas white components show sensitive clones. (B) Images of tumors consisting of 100 cells. Black cells display RAS mutant cells. Percentages on each tumor show those of RAS mutant cells, which also indicate the detection thresholds of KRAS testing being acceptable in Japan ranging from 1% to 25%. (C) Regardless of the difference in the detection limits, for example, 1% and 10%, patients determined as the KRAS mutant group (right solid rectangle) consistently showed poor prognosis as compared with those determined as the KRAS wild-type group (left solid rectangle). However, patients in the middle rectangle with the broken line were determined as having different status according to the selected assays. F-PHFA: Fluorescence Resonance Energy Transfer (FRET)-based Preferential Homoduplex Formation Assay; PCR-rSSO: Polymerase Chain Reaction Reverse-Sequence-Specific Oligonucleotide; ARMS/S: Amplification Refractory Mutation System-Scorpion Assay.
KRAS testing is an initial step in the treatment strategy for patients with metastatic colorectal cancer (mCRC). Treating patients without KRAS mutation using two biologics, such as anti-epidermal growth factor receptor (EGFR) and anti-vascular endothelial growth factor (VEGF) antibodies, achieved a median overall survival (OS) of 30 months [1, 2]. In contrast, anti-EGFR antibody was not administered to patients with KRAS mutation, resulting in a poorer median OS of 20 months [3]. KRAS status, therefore, is strongly involved in the treatment outcome of patients with mCRC, suggesting that KRAS testing is a critical initial step in the mCRC treatment strategy.
Advances in the technology to explore genomic alterations have enabled detection of rare mutant cells that are present in <0.01% of tumor cells. Increased sensitivity of KRAS testing has achieved selection of super-responders to treatment with anti- EGFR antibody but it also excludes patients who might benefit from anti-EGFR antibodies. Sub-analysis of the CRISTAL trial showed the additional effect of anti-EGFR antibodies in patients harboring minor KRAS mutations in up to 10% of tumors [4]. In order not to deprive patients of the chance to benefit from anti-EGFR antibodies, the optimal cut-off value for KRAS testing should be determined for selection of patients likely to benefit from treatment in the first-line setting. Here, we reviewed guidelines, literatures and clinical trials to elucidate whether detection threshold of KRAS testing is appropriate.
RAS Testing Recommended in Japanese Society of Medical Oncology Clinical Guidelines
The method for RAS testing recommended in the Japanese Society of Medical Oncology Clinical Guidelines [5] includes direct sequencing with manual dissection or allele-specific PCR-based methods. Direct sequencing has the advantage of detecting known and unknown gene mutations but the sensitivity is lower (10%–25%) than that of the allele-specific PCR (1%–5%). Currently, the sensitivity of RAS testing considered acceptable in Japan ranges from 1% to 25% (Table 1), but the most suitable RAS detection sensitivity or threshold remains to be determined. Using various detection thresholds may lead to differing results, including false negative, with different assays (Figure 1B).
| Direct sequencing | PCR-rSSO | ARMS/Scorpion | F-PHFA | |
|---|---|---|---|---|
| Extracorporeal diagnostic agents | MEMGEN™KRAS | The raScreen®: KRAS Mutation Kit | OncoGuideKRAS | |
| Detectable alleles of mutation | All alleles of mutation | G12S, G12C, G12R, G12D, G12V, G12A, G13D, G13C, G13R, G13D, G13V, G13A | G12S, G12C, G12R, G12D, G12V, G12A, G13D | G12S, G12C, G12R, G12D, G12V, G12A, G13D |
| Detection sensitivity | 10%-25% | 5%-10% | 1% | 10% |
PCR-rSSO:Polymerase Chain Reaction Reverse-Sequence-Specific Oligonucleotide; F-PHFA:Fluorescence Resonance Energy Transfer (FRET)Based Preferential Homoduplex Formation Assay; ARMS/Scorpion:Amplification Refractory Mutation System-Scorpion Assay.
Table 1. KRAS testing and extracorporeal diagnostic agents.
According to the Japanese Society of Medical Oncology Clinical Guidelines [5], a detection limit within 1%–10% should be considered for RAS mutation testing. These guidelines state: Various assays of RAS testing were performed in previous clinical trials. Regardless of the different detection limits between each method, the subgroup analyses of these trials consistently demonstrated that RAS status is a predictive factor for anti-EGFR antibody therapy. While the most suitable detection sensitivity remains to be determined the detection limit within 1% to 10% should be practically considered for RAS mutation testing. This wide range of detection limits yields patients with different RAS status according to the selected assays (Figure 1C). There is no mention in the guidelines of whether they should be positive or negative.
Recommended Detection Sensitivity in KRAS Testing
Bando et al. [6] conducted a retrospective analysis to compare sensitive and quality-controlled KRAS testing with direct sequencing, and to assess the impact on treatment decision making for 159 patients refractory to fluoropyrimidine, oxaliplatin and irinotecan. They were treated with cetuximab (Cmab) monotherapy or a combination of Cmab plus irinotecan. All mutations identified by direct sequencing were confirmed by the sensitive KRAS testing, amplified refractory mutation system-Scorpion assay (ARMS/Scorpion). However, 11 (7.0%) of the 70 KRAS mutations identified by ARMS/Scorpion were not detected by direct sequencing, indicating that the latter method included 7.0% of patients with false-negative results (Figure 2A).
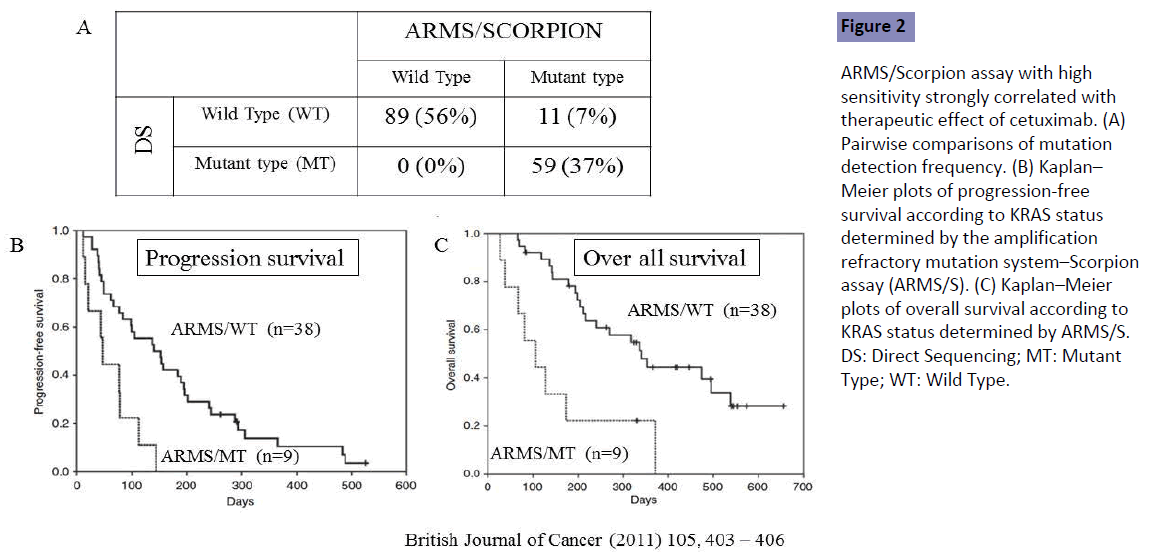
Figure 2: ARMS/Scorpion assay with high sensitivity strongly correlated with therapeutic effect of cetuximab. (A) Pairwise comparisons of mutation detection frequency. (B) Kaplan– Meier plots of progression-free survival according to KRAS status determined by the amplification refractory mutation system–Scorpion assay (ARMS/S). (C) Kaplan–Meier plots of overall survival according to KRAS status determined by ARMS/S. DS: Direct Sequencing; MT: Mutant Type; WT: Wild Type.
The measured outcomes were compared between true KRAS wild-type (WT) patients determined by both direct sequencing and ARMS/Scorpion (n=38), and false-negative patients [KRAS WT by direct sequencing but mutant (MT) by ARMS/Scorpion (n=9)]. The response rate was 16% in the ARMS/WT group but no objective response was seen in the ARMS/MT group. Poor outcomes in both progression-free survival (PFS) and OS were seen in the ARMS/MT as compared with the ARMS/WT group (Figures 2B and 2C). Direct sequencing with low sensitivity included the false-negative patients showing poor outcomes in both PFS and OS, demonstrating that ARMS/Scorpion is better than low-sensitivity DS for prediction of patient outcome. Patients in that study were refractory to previous first- and second-line fluoropyrimidine, oxaliplatin and irinotecan, indicating that detection sensitivity should be high for assessment of KRAS status in patients receiving second- or laterline treatment. Tougeron et al. [7] showed similar results for patients with mCRC with second- or later-line treatment, but no study has been conducted in the first-line setting.
What We Learned from Clinical Trials
Recent clinical trials have elucidated KRAS status using different detection methods, including Sanger sequencing, with a sensitivity of 10–20%; pyrosequencing, with a sensitivity of <5%; WAVE-based SURVEYOR Scan Kits (Transgenomic, Omaha, NE, USA), with a sensitivity of ~ 1%; and BEAMing technology (Sysmex Inostics, Baltimore, MD, USA), with a sensitivity of 0.01% (Table 2). Increased detection sensitivity revealed that the frequency of KRAS mutation was different in each method. Table 3 shows different frequencies of new RAS mutations in exons 3 and 4 in the OPUS and FIRE-3 trials. The OPUS trial exhibited increased frequencies of KRAS mutation in exons 3 and 4 as compared with the FIRE-3 trial. The OPUS trial used the high-sensitivity detection method (1%) but the FIRE-3 used the relatively low (5%). The samples examined in both studies were obtained from resected tumor specimens before chemotherapy. Increased frequencies of KRAS mutation determined by the assay with high sensitivity revealed that a small number of mutant clones existed in tumors before chemotherapy but they were not detected by the low-sensitivity detection method (Figure 3A). The low-sensitivity detection method leads to false-negative patients harboring a small number of KRAS mutant clones.
| Clinical trials | KRAS testing | Sensitivity |
|---|---|---|
| OPUS/CRISTAL | Beaming | 0.01% |
| CALGB | Beaming | 0.01% |
| PEAK | SURVEYOR | 1% |
| FIRE-3 | Pyrosequencing | 5% |
| PRIME | Sanger sequencing | 10–20% |
Table 2.Methods of KRAS testing and their sensitivities used in various clinical trials.
| Clinical trials | KRAS mutation | Sensitivity | |
|---|---|---|---|
| Exon 3 | Exon 4 | ||
| OPUS | 6.8% | 9.3% | 1% (high) |
| FIRE-3 | 4.3% | 4.9% | 5% (low) |
Table 3.Frequency of KRAS mutation in exons 3 and 4, and sensitivity of KRAS testing used in the OPUS and FIRE-3 trials.
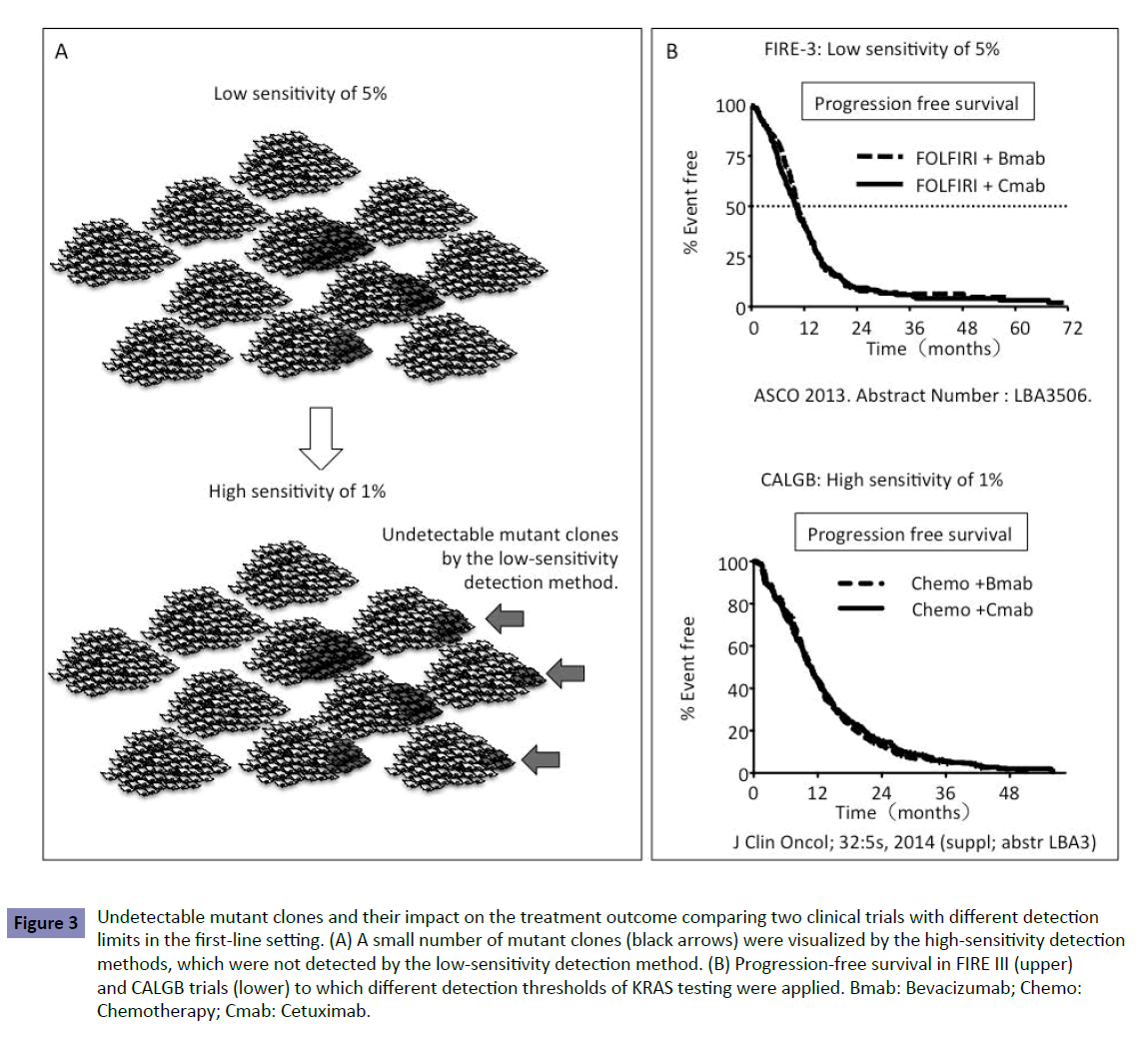
Figure 3: Undetectable mutant clones and their impact on the treatment outcome comparing two clinical trials with different detection limits in the first-line setting. (A) A small number of mutant clones (black arrows) were visualized by the high-sensitivity detection methods, which were not detected by the low-sensitivity detection method. (B) Progression-free survival in FIRE III (upper) and CALGB trials (lower) to which different detection thresholds of KRAS testing were applied. Bmab: Bevacizumab; Chemo: Chemotherapy; Cmab: Cetuximab.
Impact of Undetectable Mutant Clones on First-Line Treatment
To estimate the impact of these undetectable clones on firstline treatment with anti-EGFR antibody, we compared the outcomes in two phase III trials, FIRE-3 and CALGB80405. The FIRE-3 trial was conducted to compare FOLFIRI+Cmab with FOLFIRI+bevacizumab (Bmab) [8] and the CALGB80405 trial compared FOLFOX+Cmab with FOLFOX+Bmab [1]. The threshold of 5% in KRAS testing was applied to the FIRE-3 trial, where as 1% was applied to the CALGB80405 trial. Because of the detection limit of 5% in KRAS testing in the FIRE-3 trial, the trial included false-negative patients with a small number of KRAS mutations in up to 5% of clones in the tumor. Both trials showed no difference in PFS between patients treated with Cmab and Bmab despite the different detection limits in each trial (Figure 3B). Considering patients treated with Bmab in each trial, the results indicated that there was no difference in PFS between patients treated with Cmab in the FIRE-3 trial and CALGB80405 trial. The 1% and 5% detection thresholds did not affect PFS in the first-line setting between the CALGB80405 and FIRE-3 trials (Figure 3B). If the detection threshold of 5% had influenced PFS, patients treated with Cmab in the FIRE-3 trial would have shown poor PFS because of inclusion of false-negative patients.
Pitfalls of KRAS Testing for First-Line Treatment of Patients with mCRC
No study has shown the optimal detection threshold of KRAS testing and its effects on the outcome of first-line treatment in patients with mCRC. However, there is accumulating data to show that a detection threshold up to 10% is acceptable. RAS testing was performed by the BEAMing method (0.1% detection sensitivity) in the CRISTAL trial, which compared first-line FOLFIRI +Cmab with FOLFIRI+placebo in patients with CRC. Subgroup analysis of KRAS detection threshold in the CRISTAL trial showed that additional effects of Cmab were expected, as determined by a detection threshold up to 10% [9] (Figure 4). Patients are expected to achieve the median OS of 30 months by using both anti- EGFR and VEGF antibodies (Figure 5 upper). Increasing the KRAS detection threshold to 1% may exclude these patients may benefit from anti-EGFR antibody treatment, resulting in a shorter OS of 20% months (Figure 5 lower). The excluded patients may have accounted for 10% of the total. The optimal cutoff value of KRAS testing, therefore, should be determined by conducting a clinical trial in the first-line setting (Figure 6).
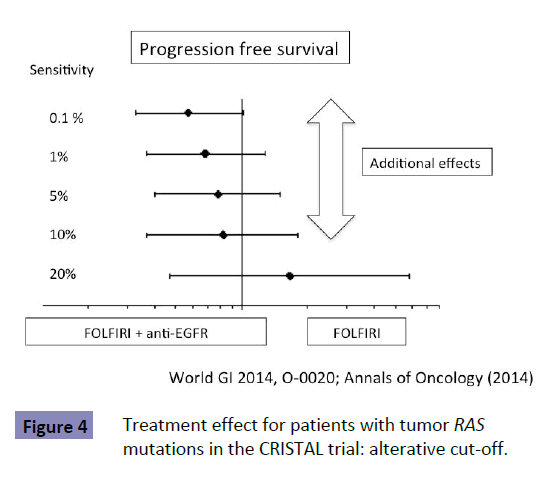
Figure 4: Treatment effect for patients with tumor RAS mutations in the CRISTAL trial: alterative cut-off.
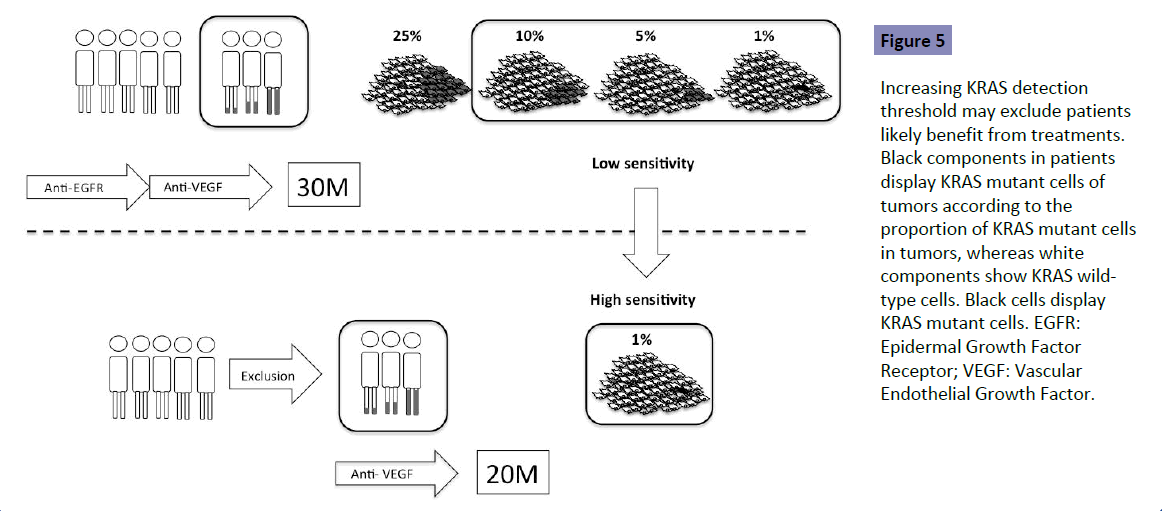
Figure 5: Increasing KRAS detection threshold may exclude patients likely benefit from treatments. Black components in patients display KRAS mutant cells of tumors according to the proportion of KRAS mutant cells in tumors, whereas white components show KRAS wildtype cells. Black cells display KRAS mutant cells. EGFR: Epidermal Growth Factor Receptor; VEGF: Vascular Endothelial Growth Factor.
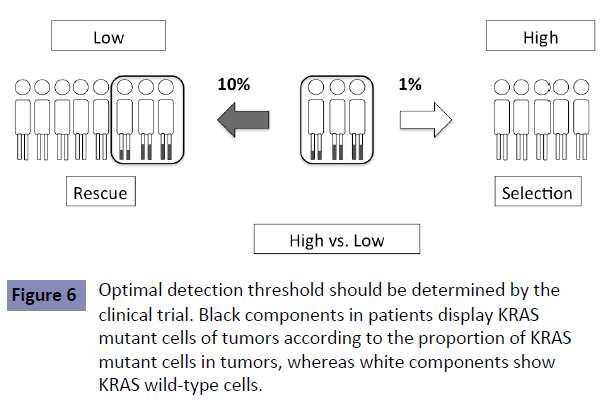
Figure 6: Optimal detection threshold should be determined by the clinical trial. Black components in patients display KRAS mutant cells of tumors according to the proportion of KRAS mutant cells in tumors, whereas white components show KRAS wild-type cells.
Impact of Undetectable Clones on Second-Line Treatment
The FIRE-3 trial showed a different outcome between patients treated with Bmab and Cmab (Figure 7A). This difference in the survival curves was seen 24 months after induction of first-line chemotherapy, which was during the second-line treatment. In the second-line setting, patients assigned to Bmab treatment in the first-line setting showed poor outcome as compared with those assigned to Cmab. Most of the patients treated with firstline treatment underwent second-line treatment. As described before, the low-sensitivity detection methods influenced the outcome of patients treated with second- or third-line Cmab [6], which is consistent with the outcome of patients treated with Cmab in the FIRE-3 trial.
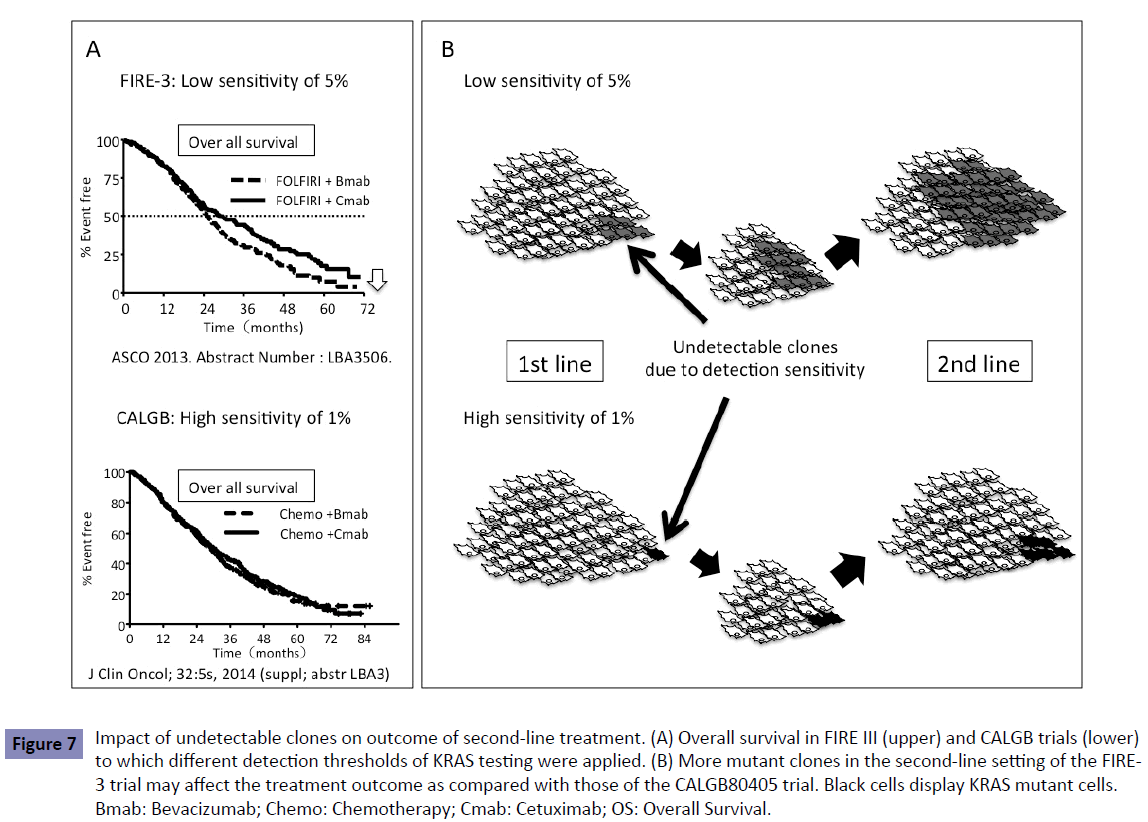
Figure 7: Impact of undetectable clones on outcome of second-line treatment. (A) Overall survival in FIRE III (upper) and CALGB trials (lower) to which different detection thresholds of KRAS testing were applied. (B) More mutant clones in the second-line setting of the FIRE- 3 trial may affect the treatment outcome as compared with those of the CALGB80405 trial. Black cells display KRAS mutant cells. Bmab: Bevacizumab; Chemo: Chemotherapy; Cmab: Cetuximab; OS: Overall Survival.
Change in Genomic Profiles Determined by Liquid Biopsy and its Influence on Treatment
A blood-based technology platform tracking circulating tumor DNA, known as liquid biopsy, allows multiple testing over time, monitoring real-time changes within the tumor and evaluation of therapeutic response. Liquid biopsy was achieved using the BEAMing technology or digital PCR with a high sensitivity of 0.1%–0.01%. Liquid biopsy shows the dynamic change in genomic profiles such as KRAS status during treatment. We and other groups [10-12] have reported that patients with KRAS WT in the tumor display the KRAS mutant clones in the blood during treatment (Figure 8). This suggests that latent undetectable KRAS mutant clones in the primary tumor increase and appear in the blood during treatment. KRAS mutant clones may be replaced in the treated tumor that originally consisted of KRAS WT clones. This could happen when more undetectable KRAS mutant clones are present in the primary tumor, determined by a low-sensitivity detection limit, resulting in resistance to second- and third-line anti-EGFR antibody. This is one explanation of why patients treated with second-line Cmab in the FIRE-3 trial line showed poor outcome (Figure 7B).
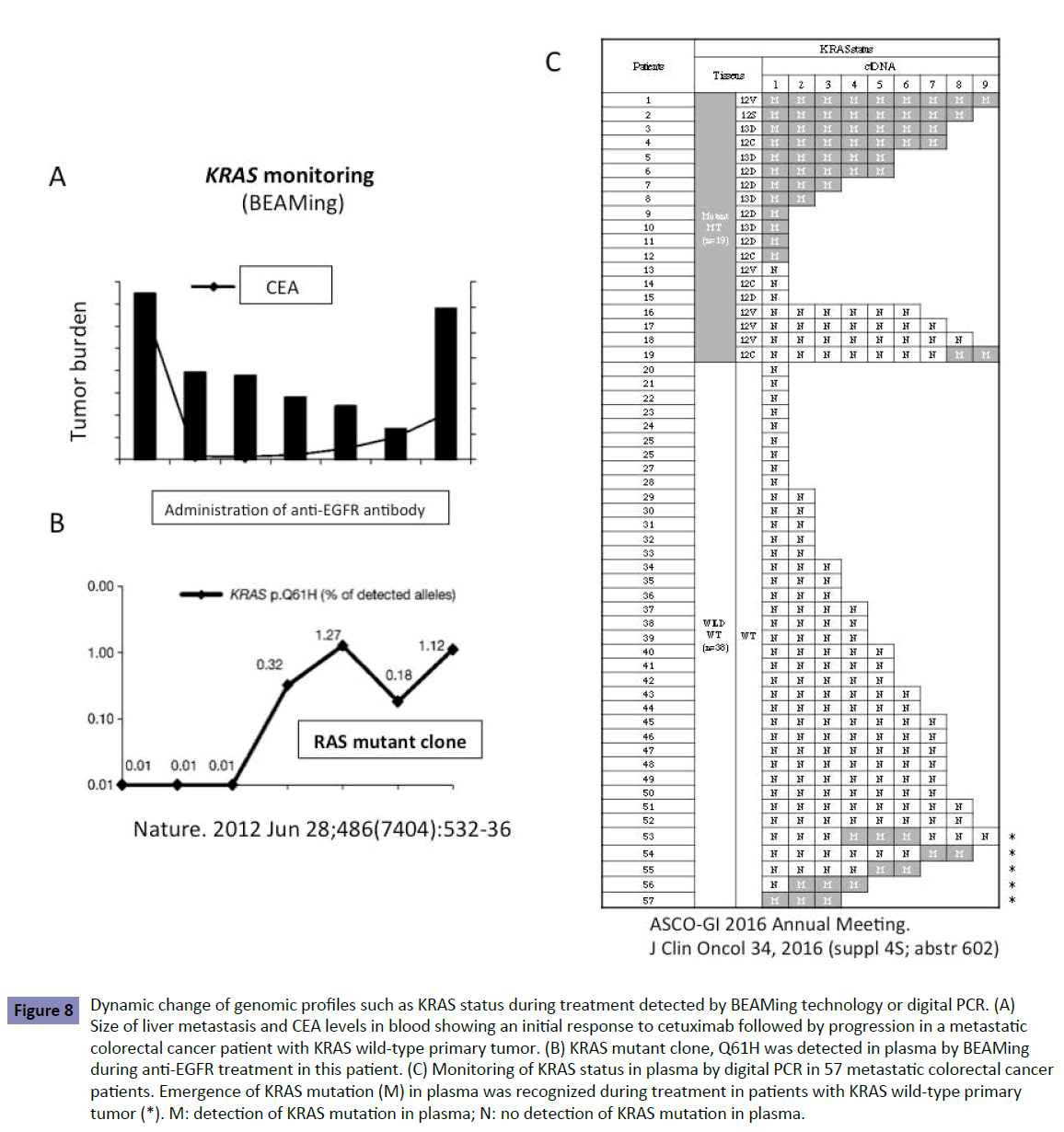
Figure 8: Dynamic change of genomic profiles such as KRAS status during treatment detected by BEAMing technology or digital PCR. (A) Size of liver metastasis and CEA levels in blood showing an initial response to cetuximab followed by progression in a metastatic colorectal cancer patient with KRAS wild-type primary tumor. (B) KRAS mutant clone, Q61H was detected in plasma by BEAMing during anti-EGFR treatment in this patient. (C) Monitoring of KRAS status in plasma by digital PCR in 57 metastatic colorectal cancer patients. Emergence of KRAS mutation (M) in plasma was recognized during treatment in patients with KRAS wild-type primary tumor (*). M: detection of KRAS mutation in plasma; N: no detection of KRAS mutation in plasma.
The dynamic change of genomic profiles including KRAS status, therefore, should be monitored by liquid biopsy to enable the optimal choice of drugs during treatment. The monitoring of KRAS testing in one recent study [13] showed that KRAS mutant clones decreased in blood during treatment with anti-EGFR antibody. This suggests that sensitivity to anti-EGFR antibody could be recovered and re-introduction of treatment could be effective (Figure 9).
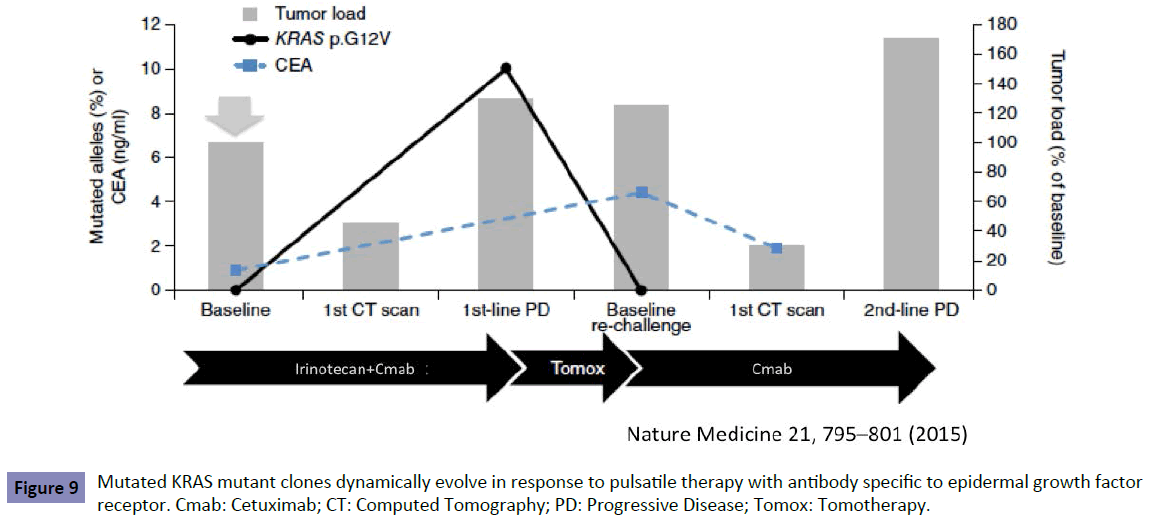
Figure 9: Mutated KRAS mutant clones dynamically evolve in response to pulsatile therapy with antibody specific to epidermal growth factor receptor. Cmab: Cetuximab; CT: Computed Tomography; PD: Progressive Disease; Tomox: Tomotherapy.
What the Forthcoming Clinical Trial will Teach Us?
In this article, we have shown that there is no difference in the influence of detection thresholds on the outcome of first-line treatment, despite the difference in the second-line setting. The question is what is the optimal detection threshold in sequential treatments from first- to second-line setting? It should be determined by clinical trial but two scenarios must be considered. In case that a high-sensitivity detection threshold, for example 1%, is appropriate by the forthcoming trial for selection of patients likely to benefit from first-line treatment with anti-EGFR antibody, false-negative patients determined by the low-sensitivity detection threshold would be excluded. Any sequential treatments would be acceptable from first- to second-line setting, however, the detection threshold in all the RAS testing widely accepted, including RASKET, should be changed to high sensitivity (Figure 10A). In case that a lowsensitivity detection threshold, for example 10%, is appropriate, false-negative patients would be rescued. However, we should consider the treatment sequence, such that anti-EGFR antibody precedes anti-VEGF antibody because anti-EGFR treatment in the second-line settin showed poor outcome (Figure 10B) as shown before (Figure 2).
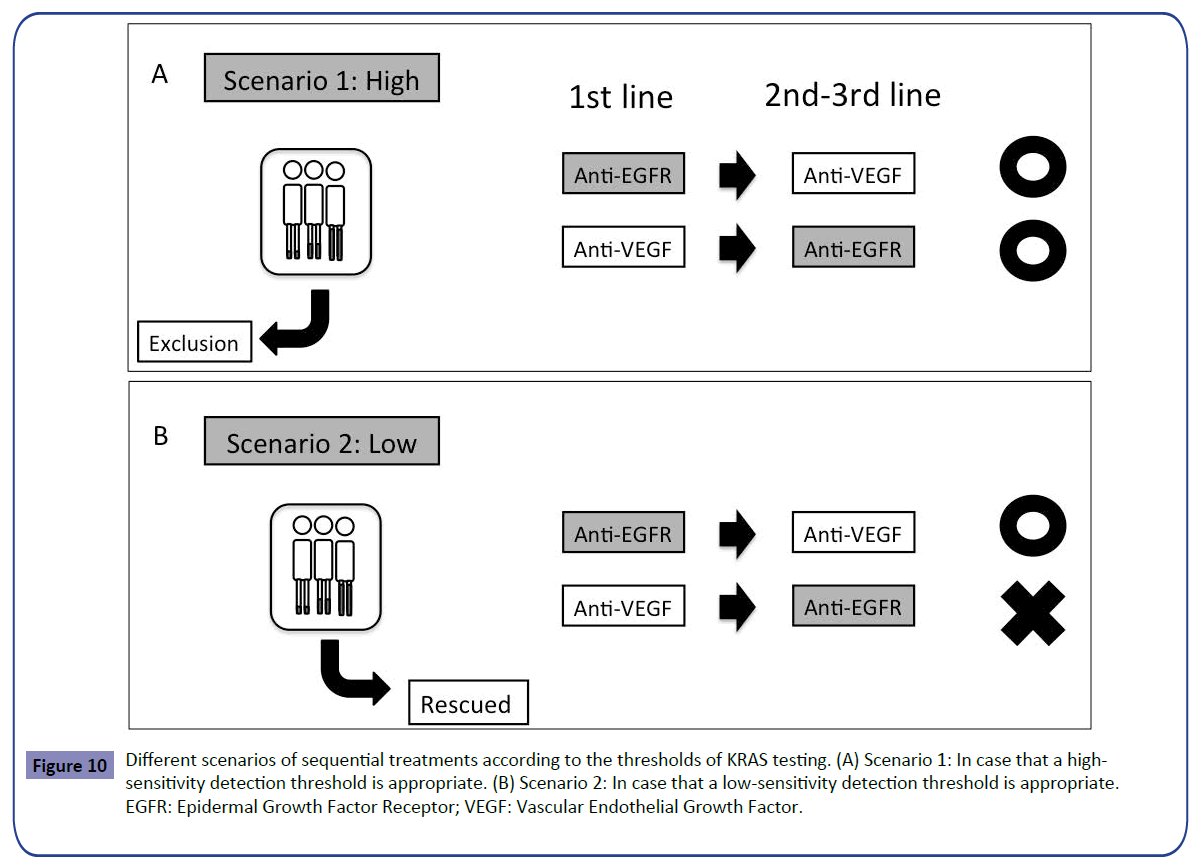
Figure 10: Different scenarios of sequential treatments according to the thresholds of KRAS testing. (A) Scenario 1: In case that a highsensitivity detection threshold is appropriate. (B) Scenario 2: In case that a low-sensitivity detection threshold is appropriate. EGFR: Epidermal Growth Factor Receptor; VEGF: Vascular Endothelial Growth Factor.
Conclusion
In conclusion, the optimal cut-off value of KRAS testing was not determined for selection of patients likely to benefit from firstline treatment with anti-EGFR antibody. Thus, we recommend that a clinical trial should be conducted to compensate for the pitfalls of KRAS testing.
Conflict of Interest
The authors declare that they have no conflict of interest.