Research Article - (2018) Volume 4, Issue 1
Identification of Leachables from Trametes versicolor in Biodegradation Experiments
Åke Stenholm1,2*, Mikael Hedeland1,3, Torbjörn Arvidsson1,4 and Curt E Pettersson1
1Department of Medicinal Chemistry, Uppsala University, BMC Box 574, SE-751 23, Uppsala, Sweden
2GE Healthcare Bio-Sciences AB, Björkgatan 30, SE-751 84 Uppsala, Sweden
3National Veterinary Institute (SVA), Department of Chemistry, Environment and Feed Hygiene, SE-751 89, Uppsala, Sweden
4Medical Products Agency, Box 26, SE-751 03, Uppsala, Sweden
*Corresponding Author:
Åke Stenholm
Department of Medicinal Chemistry,
Uppsala University, BMC Box 574, SE-751
23, Uppsala, Sweden
E-mail: ake.stenholm@ilk.uu.se
Received date: March 14, 2016; Accepted date: April 19, 2016; Published date: April 22, 2016
Citation: Stenholm A, Hedeland M, Arvidsson T, Pettersson CE (2018) Identification of Leachables from Trametes versicolor in Biodegradation Experiments. Vol. 4 No. 1:1
The transport of fungal-derived compounds from Trametes versicolor to the environment was investigated. Fatty acids and sphingoids were identified at the outlet of a bioreactor containing an acidic nutrient solution and immobilized fungal mycelia. The analyses were conducted using UHPLC-Q-TOF-MS (/MS). Eleven fatty acids, including C20:0, C18:1-OH and C20:0-OH that have not been previously described for this species, were detected. The identities of myristic acid (C14:0), palmitic acid (C16:0) and stearic acid (C18:0) were confirmed using reference standards. Six sphingoids, including Sph (t18:0), Sph (t18:1), Sph (d18:0), Sph (d18:1), Sph (d16:0) and Sph (d16:1), were tentatively identified, and the identities of Sph (d18:0) and Sph (d18:1) were confirmed by reference standards. The findings show that an array of compounds, with concentrations at the μgL-1 level, was easily transported from the fungal mycelia. This is of concern when the investigated species is used in biodegradation experiments of xenobiotics and conclusions are to be drawn on the quality of the treated water. The study thus shows that the chemical composition of water treated with Trametes versicolor is also influenced by the immobilized fungus itself. The lipids that were detected, including fatty acids and sphingoids do not present any threat to the environment since they are not toxic. At μgL-1 concentration levels, they are soluble in water.
Keywords
Xenobiotics; Immobilized fungus; Sphingoids; lipids
Introduction
The enzyme-catalysed biodegradation of environmentally hazardous compounds in lab-scale experiments is well documented, including research that has studied the biodegradation potential of extracellular enzymes secreted by white rot fungi (WRF) mycelia [1].The particular interest in these fungal species is related to their effective biodegradation of recalcitrant pharmacologically active compounds including endocrine disrupting compounds (EDCs) [2,3]. These studies typically emphasize three objectives: (A) removal of target compounds; (B) correlation of this removal to measured enzymatic activities; and (C) identification of degradation products, including toxicity assessments [4-6]. However, there is limited knowledge regarding which constituents are leached from the mycelial cells into the medium. Secretion and excretion are two concepts that are commonly used to describe the processes through which compounds penetrate cell walls. Secretion is the active process of releasing and transporting chemical substances, like extracellular enzymes (exoenzymes), out of a cell, while excretion is the passive transport of waste products that have no further utility. Active processes do not only comprise the secretion of exoenzymes (exocytosis) from fungal cells, but can include endocytosis, in which extracellular macromolecules are engulfed by the cell membrane and imported into the cell [7]. Exocytosis describes a process in which membrane-bound vesicles containing enzymes, toxins and lipids -fuse with the plasma membrane in hyphae [8]. Like the plasma membrane, the vesicle membranes also contain lipid bilayers. Upon fusing, the vesicle membrane proteins and lipids move to the outer region of the fungal plasma membrane via the Spitzenkörper, i.e. a vesicle supply centre present in hyphae.
A passive transport of molecules out from the fungal cell is accomplished through the cell wall pores providing that the molecules are not too large. However, permeability of up to 270 kDa dextran molecules have been reported [9]. It must be emphasized that the cell wall is permeable in both directions, and thus also includes the influx of nutrients, water and ions [10]. A fungal hypha tip, including cell organelles and simplified transport mechanisms, is illustrated in Figure 1. Thus, lipids from vesicles, as well as passively transported compounds originating from cell membranes and the cytosol, can be expected in biodegradation experiments. The mild conditions that are used in biodegradation experiments cannot be compared with the conditions utilized during harsh mycelium extraction methodologies. The fungal cell wall, which has previously been described as carbohydrate armour [11], contains chitin, glucan polysaccharides and mannoproteins [9]. T. versicolor mycelia are extracted with hot water (100oC) to obtain the polysaccharide Krestin (PSK) and polysaccharopeptides (PSPs), both of which are used to supplement cancer chemo- and radiotherapy [12]. The boiling water is used to weaken the cell walls, as high temperatures are necessary for successful cell wall lysis. A hot alkaline solution can also be used for this purpose [11]. Lipids, alkaloids and phenolic compounds can be extracted from disrupted fungal mycelia with organic solvents [13-16]. In these studies, the cell walls were initially mechanically crushed, but lipids can be extracted from mycelial cells prior to any milling procedure.
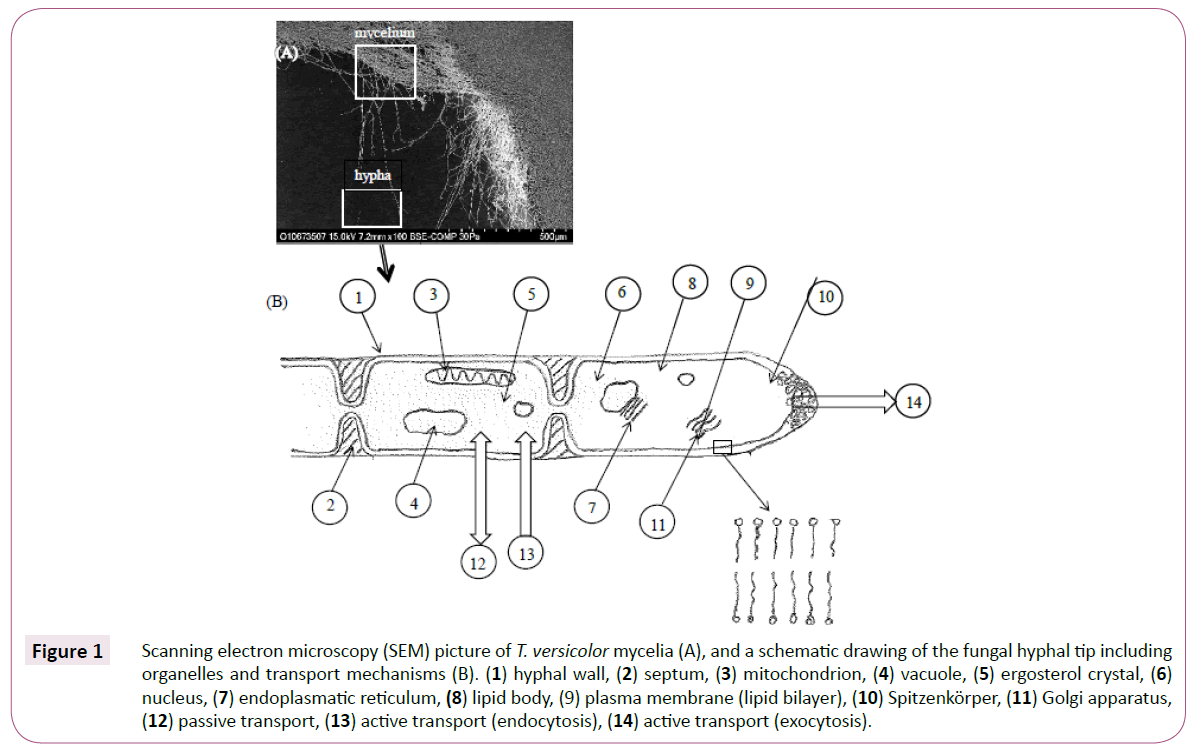
Figure 1: Scanning electron microscopy (SEM) picture of T. versicolor mycelia (A), and a schematic drawing of the fungal hyphal tip including organelles and transport mechanisms (B). (1) hyphal wall, (2) septum, (3) mitochondrion, (4) vacuole, (5) ergosterol crystal, (6) nucleus, (7) endoplasmatic reticulum, (8) lipid body, (9) plasma membrane (lipid bilayer), (10) Spitzenkörper, (11) Golgi apparatus, (12) passive transport, (13) active transport (endocytosis), (14) active transport (exocytosis).
In the present investigation, the mycelia were grown in a slightly acidic nutrient solution thereby maintaining the cell wall structure. The study aimed to identify leachables from T. versicolor mycelia, and applied a previously described experimental procedure that was used in the investigation of the biodegradation and adsorption mechanisms of the NSAID diclofenac [17]. Bioreactor experiments were performed at pH 4, which was chosen specifically to minimize bacterial growth, under non-sterile conditions. In addition to fungal mycelia, the reactor contained a nutrient solution consisting of glucose and ammonium tartrate dissolved in tap water. Fresh nutrient solution containing diclofenac was pumped through the bioreactor during the experiments. The search for T. versicolor leachables presented in this paper was performed on an experimental culture containing living fungal mycelia. UHPLC-Q-TOF MS and MS/MS were used to identify the compounds. The identity of fungal derived leachables resulting from T. versicolor mediated biodegradation has previously not been studied, and the presented research adds valuable information about the chemical composition of fungaltreated water, specifically, that fungal constituent as well as the biodegradation products of pharmacologically active compounds (PhACs) and EDCs can be present in treated wastewaters.
Materials and Methods
Fungus
T. versicolor (strain AG1383) was obtained from PhD Ivana Eichlerová at the Institute of Microbiology, the Culture Collection of Basidiomycetes, Institute of Microbiology, and Academy of Sciences of the Czech Republic in Prague. The fungus was maintained by subculturing every second month onto disks containing modified malt extract agar supplied by the National Veterinary Institute (SVA, Uppsala, Sweden) at ambient temperature.
General experimental procedures
The nutrient solution used throughout the bioreactor experiments (including fungal growth onto immobilization supports and biodegradation experiments) contained 6.0 g L-1 D-(+)-glucose (≥ 99.5%) and 3.3 g L-1 ammonium tartrate (≥ 98%) with a carbon: nitrogen molar ratio (C:N) of 6.6. The pH of the solution was adjusted to 4.0 with 1.0 M LiChrosolv®sulfuric acid (Merck, Darmstadt, Germany). Glucose, ammonium tartrate, DLDihydrosphingosine, synthetic (≥ 98%), D-Sphingosine (≥ 98%), myristic acid (≥ 99, 5%), palmitic acid (≥ 99%) and stearic acid (≥ 98.5%), were purchased from Sigma-Aldrich (St. Louis, MO). Diclofenac sodium salt (≥ 98%), was purchased from Sigma- Aldrich, (St. Louis, MO). Diclofenac-d4 with an isotopic enrichment of 98 atom% deuterium was obtained from C/D/N Isotopes Inc. (Quebec, Canada). Softened industrial tap water (removed calcium and magnesium) from General Electric Healthcare, Uppsala, Sweden was used unless otherwise specified. The mobile phases that were used for the UHPLC-Q-TOF MS and MS/ MS analyses were LiChrosolv® Hypergrade acetonitrile (≥ 99.9%) and LiChrosolv® water for chromatography. The chemicals were purchased from Merck (Darmstadt, Germany). Ammonium acetate (≥ 99.0%) for the preparation of the mobile phase was obtained from Fluka (Darmstadt, Germany). The immobilization supports (carriers), which contained polyurethane foam (PUF) were purchased from Eheim GmbH & Co. KG (Deizisau, Germany). The model type was Eheim pickup 160.
Preparation of reference solutions of fatty acids and sphingoids used for confirmation of chemical structures
Reference solutions of myristic (C14:0), palmitic (C16:0) and stearic acid (C18:0) were prepared at 100 μgL-1 concentrations using the nutrient solution as a solvent. Stock solutions with a concentration of 5 mgL-1 were first done to allow the acids, stearic acid in particular, to dissolve. Additionally, reference sphingoid solutions were prepared. The chosen compounds were Sph (d18:0) (dihydrosphingosine), and Sph (d18:1) (sphingosine). They were dissolved to 100 μgL-1 concentrations in nutrient solution. Analyses of the reference solutions were performed within 24 h.
UHPLC-Q-TOF analysis
An Agilent Technologies 6550 funnel Q-TOF LC/MS system (Agilent Technologies, Santa Clara, CA) including an Agilent Technologies 1290 Infinity UHPLC system consisting of a 1260 iso pump (G1310B), 1290 binary pump (G4220A), thermostat (G1330B), 1290 sampler (G4226A), 1290 thermostated column compartment (G1316C) and electrospray ionization (ESI) was used for the UHPLC-Q-TOF MS and MS/MS analyses. Agilent Mass Hunter software (version. 06.00) was used for data acquisition and processing. The drying gas flow rate and temperature were set to 14 Lmin-1 and 150oC, respectively, while the sheath gas flow and temperature were set to 11 Lmin-1 and 350oC, respectively. The capillary voltage was set to 4000 V. Positive and negative polarities were assessed, with the acquisition rate set to 3 scan sec-1. The scanned m/z range was 100-1000. Lock masses with m/z 121.05087300 (protonated purine) and m/z 922.00979800 (protonated hexakis-(1H, 1H, 3H-tetrafluoropentoxy) phosphazene) were used during positive ionization mode analyses, whereas m/z 112.9856 (TFA anion) and m/z 980.0164 (hexakis-(1H,1H,3H-tetrafluoro-pentoxy) phosphazene) were chosen for the negative ionization mode analyses. In MS/ MS, the collision-induced dissociation (CID) energies were set to 10, 20 and 40 V.
Before each analysis occasion (i.e day), the instrument was calibrated in positive and negative modes using the Agilent Check Tune procedure, which demands a mass accuracy <1 ppm and a resolution >20000. The presence of lock masses was confirmed during each analysis sequence. A Zorbax Eclipse plus C18 rapid resolution HD (2.1 x 50 mm, 1.8 μm) UHPLC-column from Agilent Technologies was used for separation purposes. The flow rate and temperature were set to 0.3 mLmin-1 and 30oC, respectively. Mobile phases A and B consisted of Lichrosolv water including 2.0 mM ammonium acetate and hypergrade acetonitrile, respectively. Mobile phase A had a pH of 7.0. The composition of the mobile phase consisted initially of 90% A which was then subjected to a linear decrease to 10% .A over six minutes, followed by an isocratic period of one minute. The concentration of the mobile phase was then linearly increased from 10% A to 90% .A over a time period of one minute to equilibrate the column. The injection volumes were 5 μL.
Inoculation and bioreactor experiments
A Biostat® B reactor (B. Braun Biotech International, Melsungen, Germany) containing a 2 L reaction vessel and equipped with an effluent Pump P-1 peristaltic pump (Pharmacia Biotech, Uppsala, Sweden) was used for bioreactor experiments aiming to remove the non-steroidal anti-inflammatory drug diclofenac (Figure 2) [17]. The reactor was run using a semi-continuous supply of nutrients and diclofenac and a constant peristaltic pump-mediated effluent flow of 1 mLmin-1 (hydraulic retention time (HRT) of 33 h). Fresh nutrient solution containing diclofenac (10 mgL-1) was added to the reactor via the substrate supply port (port 6 in Figure 2) when the liquid level in the reactor reached the determined minimum level (at the lower end of the liquidlevel sensor, shown in port 4 in Figure 2). New solution was added until the total volume reached 2 L. The fungal-treated solution was pumped out of the reactor through a titanium filter in the harvesting pipe (port 1 in Figure 2) to exclude any mycelia residues from the collected samples. The pH was kept at 4.0 with an automatic acid and base doser (1.0 M NaOH and sulfuric acid) [17].
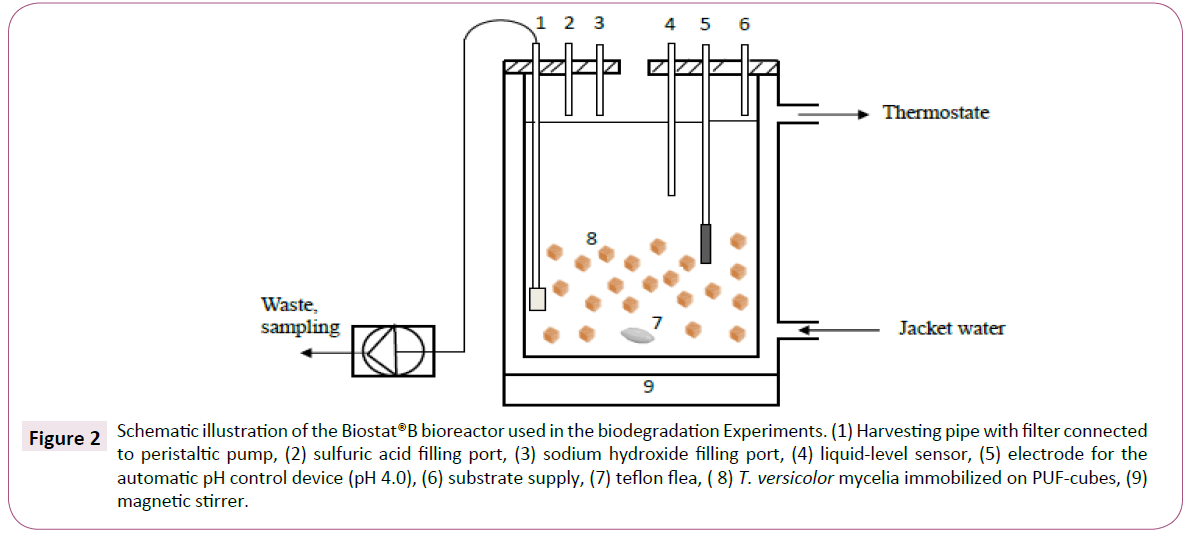
Figure 2: Schematic illustration of the Biostat®B bioreactor used in the biodegradation Experiments. (1) Harvesting pipe with filter connected to peristaltic pump, (2) sulfuric acid filling port, (3) sodium hydroxide filling port, (4) liquid-level sensor, (5) electrode for the automatic pH control device (pH 4.0), (6) substrate supply, (7) teflon flea, ( 8) T. versicolor mycelia immobilized on PUF-cubes, (9) magnetic stirrer.
A total of 14.2 g of PUF pieces, each with an approximate 1 cm3 cubic size (cut with scissors) were added to the bioreactor, followed by the addition of 2 L of nutrient solution. The vessel was then autoclaved at 125oC for 30 min, after which 35 agar plugs containing mycelia were added. The fungus was allowed to grow in the bioreactor under non-flowing conditions for one week. Thereafter, nutrient solution containing diclofenac (10 mg L-1) was fed into the reactor under flowing conditions (port 6 in Figure 2). Samples, 4 mL each, for the UHPLC-Q-TOF analyses were collected at 0.5, 1, 2, 4, 6, 22, 29, 46, 70, 94 and 166 hours through a 2 μm titan frit that was mounted onto the effluent tubing. The reactor turn-over volume was 33 hours (HRT). Thus, at the end of the experiment (166 h), 5 reactor volumes were used.
Results
In all experiments, diclofenac d-4 was present at a concentration of 100 μgL-1.The detected compounds were present at approximate μgL-1 levels. All substances were detected using positive mode ESI MS. For fatty acids (FAs), negative mode ESI (MS/MS) was used to confirm the chemical structures of the most abundant FAs.
Fatty acid composition
Eleven FAs were detected as ammonium adducts ions using positive mode ESI. Nine were tentatively determined according to their accurate masses including mass errors <2.5 ppm. Palmitic acid (16:0) and stearic acid (18:0) were the most abundant FAs identified (Table 1).
| FA | Elemental | Accurate | Exact | Mass error | tR (min) |
|---|---|---|---|---|---|
| composition | mass, m/z | mass, m/z | mDa ppm | ||
| C14:0* | C14H28O2 | 246.2431 | 246.2433 | 0.2 0.8 | 3.98 |
| C14:1 | C14H26O2 | 244.2276 | 244.2277 | 0.1 0.4 | 4.77 |
| C16:0* | C16H32O2 | 274.2746 | 274.2746 | 0.0 0.0 | 4.95 |
| C16:1 | C16H30O2 | 272.2586 | 272.259 | 0.4 1.5 | 4.42 |
| C16:1-OH | C16H30O3 | 288.2534 | 288.2539 | 0.5 1.7 | 4.63 |
| C17:0 | C17H34O2 | 288.2897 | 288.2903 | 0.6 2.1 | 5.51 |
| C18:0* | C18H36O2 | 302.3056 | 302.3059 | 0.3 1.0 | 6.14 |
| C18:1 | C18H34O4 | 300.2899 | 300.2903 | 0.4 1.3 | 5.89 |
| C18:1-OH | C18H34O3 | 316.2849 | 316.2852 | 0.3 0.9 | 4.11 |
| C18:2 | C18H32O2 | 298.2743 | 298.2746 | 0.3 0.9 | 4.84 |
| C20:0-OH | C20H40O3 | 346.3314 | 346.3321 | 0.7 2.0 | 4.58 |
*Indentity confirmed by reference standards
Table 1: Summary of the accurate mass measurements of FAs, as determined for their ammonium adduct ions using UHPLC-Q-TOF MS. The elemental compositions are shown in their uncharged state. Data for detected ions correspond to acquisitions obtained in full-scan mode.
Negative mode ESI was used to confirm the presence of C16:0 and C18:0 based on their accurate masses and retention times. Negative ion mode scans yielded higher abundances in MS mode and less noisy spectra. MS/MS was performed at three different collision-induced dissociation (CID) energies (10, 20 and 40 V). The precursor ions of C16:0 and C18:0 were present as [M-H] ionic species. For C16:0, a major ion at m/z 255.2323 was found in full-scan mode. The accurate mass corresponded well (i.e. mass error of 2.3 ppm) with the molecular formula C16H32O2 (Table 1). As a result, this was selected as a precursor ion. No major product ions could be observed in MS/MS independent of CID-energy. Similarly, no major product ions of 18:0 were found in the corresponding product ion spectra. The precursor ion of choice for C18:0 was m/z 283.2638, which corresponds to the elemental composition C18H36O2 (Table 1). The mass error was 1.4 ppm. The three fatty acid reference solutions (see Materials and Methods) were injected and, under negative ion mode, the accurate masses of the [M-H] ionic species were determined to be 227.2013 (14:0), 255.2326 (16:0) and 283.2634 (18:0), respectively. The corresponding mass errors were 0.9, - 0.8 and 1.1 ppm, respectively. The retention times differed from those presented in Table 1 by -1.3% (14:0), -1.7% (16:0) and -1.0% (18:0). The accurate mass and retention time determinations confirmed the identity of the three fatty acids. It was supported by the dominance of the parent [M-H] peaks and the absence of product ions in MS/MS spectra [18].
Sphingoid composition
Six sphingoids were tentatively identified through MS and MS/ MS using positive ionization. The chemical structures of these compounds are shown in Figure 3. All sphingoids were identified in full-scan mode as their [M + H]+ ion species. A summary of the accurate mass determinations, including mass errors and retention times, from the full-scan MS is presented in Table 2. All six sphingoids obeyed the nitrogen rule (proton adducts with even m/z).
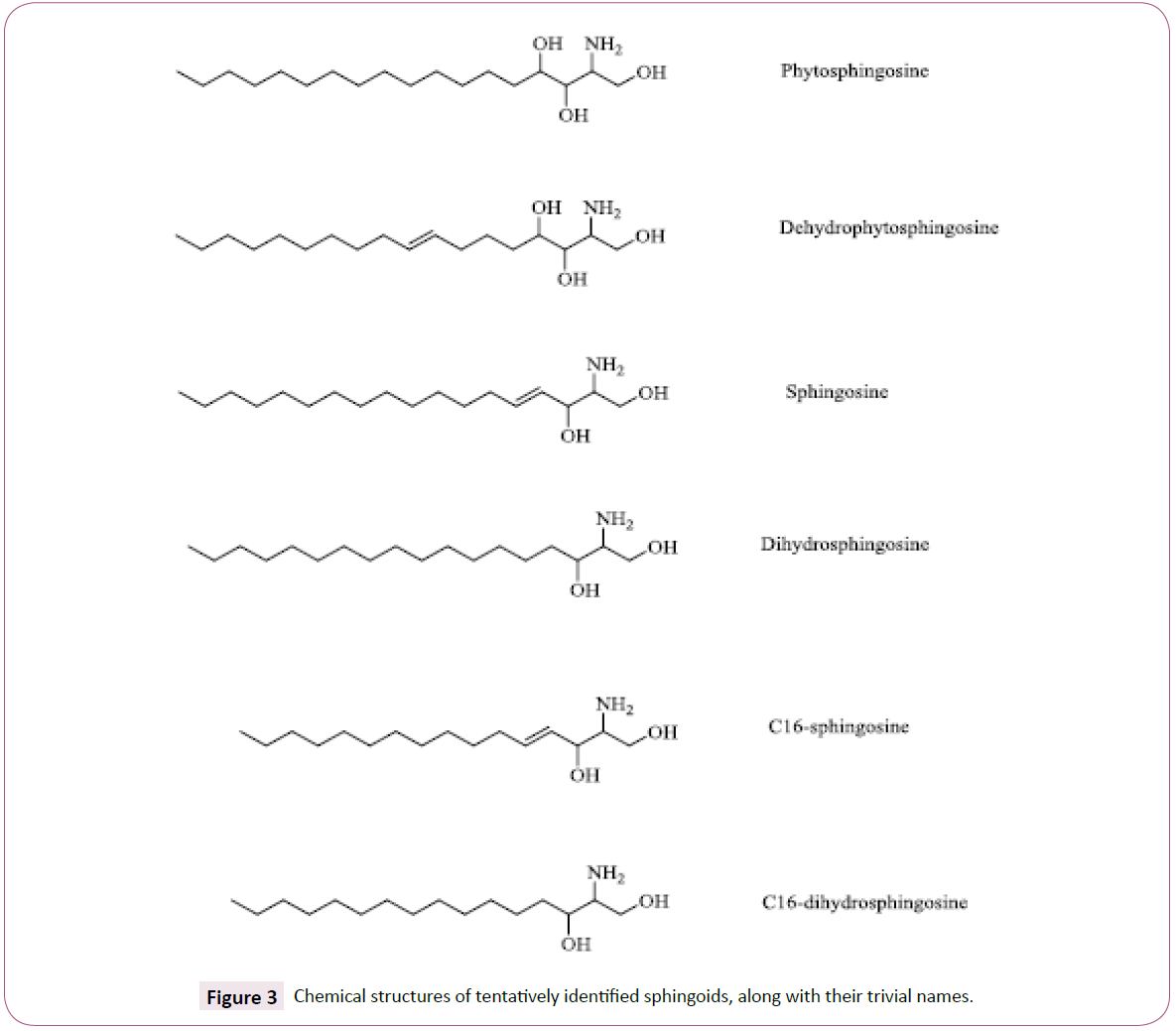
Figure 3: Chemical structures of tentatively identified sphingoids, along with their trivial names.
| Id | Trivial | Elemental | Accurate | Exact | Mass error | tR, min |
|---|---|---|---|---|---|---|
| Name | Composition | Mass (m/z) | Mass | mDa ppm | ||
| Sph (t18:0) | Phytosphingosine | C18H39NO3 | 318.3007 | 318.3003 | - 0.4 -1.3 | 3.9 |
| Sph (t18:1) | Dehydrophytosphingosine | C18H37NO3 | 316.2849 | 316.2846 | -0.3 -0.9 | 3.7 |
| Sph (d18:0)* | Dihydrosphingosine | C18H39NO2 | 302.3044 | 302.3050 | 0.6 2.0 | 6.1 |
| Sph(d18.1)* | Sphingosine | C18H37NO2 | 300.2905 | 300.2900 | -0.5 -1.7 | 5.9 |
| Sph(d16:0) | C16 dihydrosphingosine | C16H35NO2 | 274.2745 | 274.2741 | -0.4 -1.5 | 4.9 |
| Sph(d16:1) | C16 sphingosine | C16H33NO2 | 272.2590 | 272.2584 | -0.6 -2.6 | 4.7 |
d stands for dihydroxyls (di) and t for trihydroxyls (tri)
Table 2: Summary of the accurate mass measurements of sphingoids, as determined by UHPLC-Q-TOF MS. Data for detected [M + H]+ ions correspond to acquisitions obtained in full-scan mode. The elemental compositions are shown in their uncharged state.
In MS/MS, the product ion spectra showed similarities when accounting for water loss, which was most abundant at a CID of 20V. The number of water molecules lost, the m/z of the product ions and the [M + H]+ precursor ions for each sphingoid determined the chemical structures. Additionally, typical aliphatic chain fragmentation patterns were present for all compounds, mainly at CID 40V. These fragmentation patterns are argued to be of restricted use to determine the exact positions of the unsaturated carbons [19]. The product ions of Sph (t18:0), Sph (t18:1), Sph (d18:1) and Sph (d16:1) had m/z values of 44, 55, 67, 81, 95, 109, 123 and 129. However, fragments with m/z 69, 83 and 97 rather m/z 67, 81 and 95 were most abundant for Sph (d16:0) and Sph (d18:0). For the investigated sphingoids, it is assumed that a trans-double bond is present at position 4 [20], whereas the trans-double bond is present at position 8 in dehydrophytosphingosine (Sph (t18:1)) [21]. Interestingly, by examining the product ions for the sphingosines (which contain two -OH groups) in particular, the m/z 67, 81 and 95 ions indicated the presence of an unsaturation, while the m/z 69, 83 and 97 ions were significant for the saturated sphingosines.
The product ions at m/z 44.0491 and 55.0544 can be traced to C2H6N and C4H7 fragments respectively by matching the accurate masses against theoretical monoisotopic masses for known fragment ions including C, H, N and O. The mass errors were less than 10 ppm for both product ions. In Figure 4, a proposed fragmentation scheme is shown for Sph (d18:1) and Sph (d18:0). This scheme includes product ions with mass errors <10 ppm.
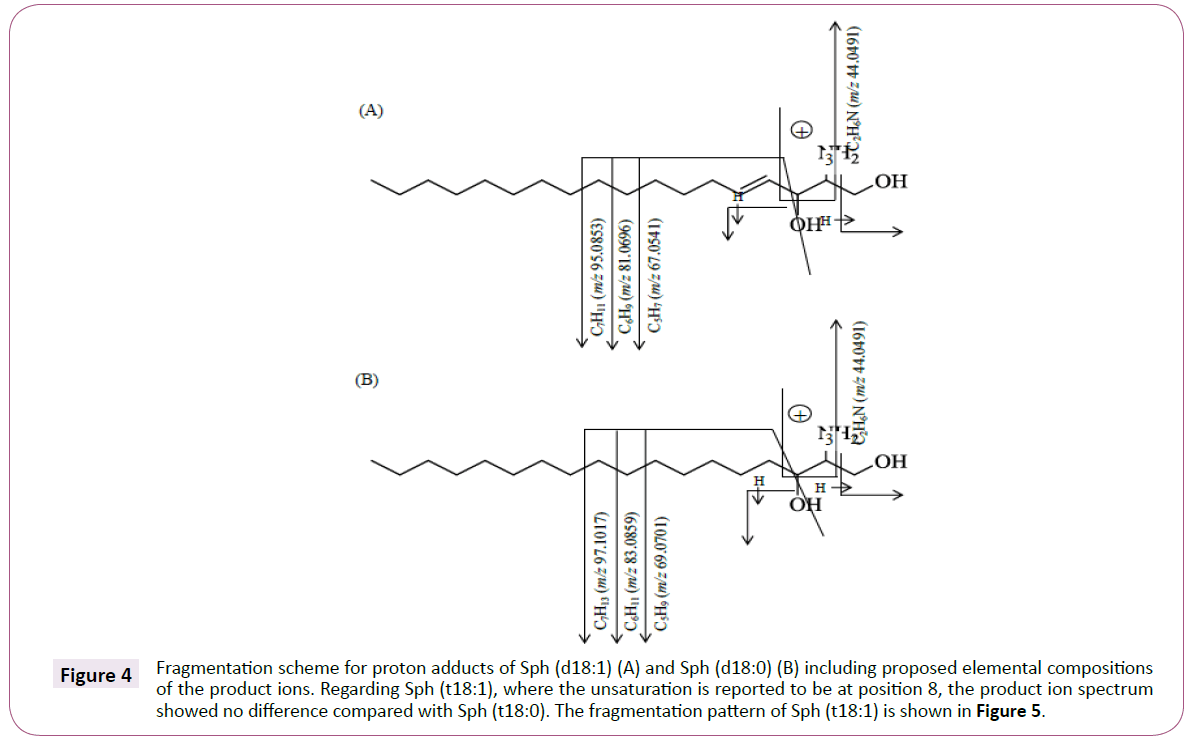
Figure 4: Fragmentation scheme for proton adducts of Sph (d18:1) (A) and Sph (d18:0) (B) including proposed elemental compositions of the product ions. Regarding Sph (t18:1), where the unsaturation is reported to be at position 8, the product ion spectrum showed no difference compared with Sph (t18:0). The fragmentation pattern of Sph (t18:1) is shown in Figure 5.
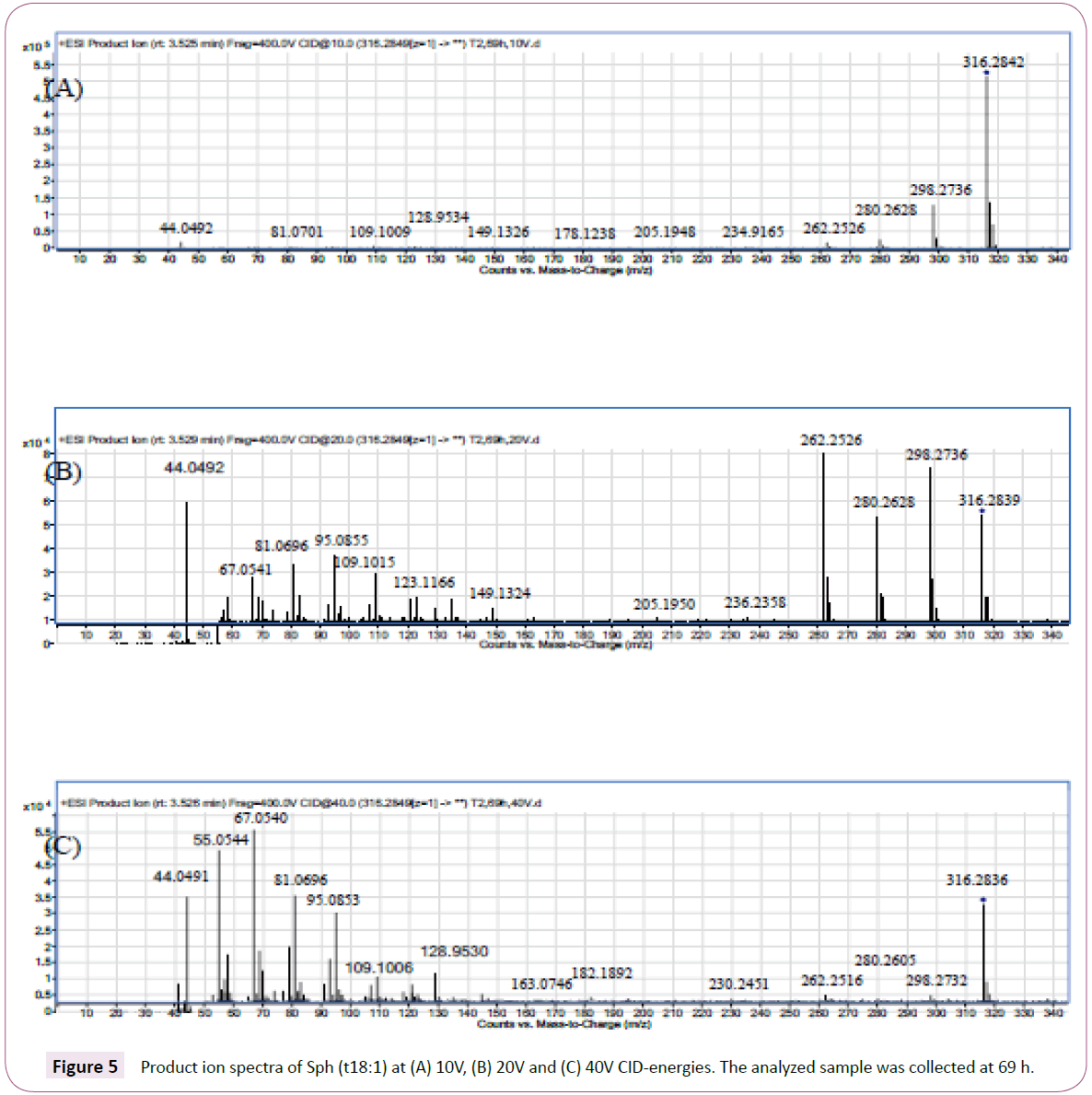
Figure 5: Product ion spectra of Sph (t18:1) at (A) 10V, (B) 20V and (C) 40V CID-energies. The analyzed sample was collected at 69 h.
The product ions at m/z 44.0491 and 55.0544 can be traced to C2H6N and C4H7 fragments respectively by matching the accurate masses against theoretical monoisotopic masses for known fragment ions including C, H, N and O. The mass errors were less than 10 ppm for both product ions. In Figure 4, a proposed fragmentation scheme is shown for Sph (d18:1) and Sph (d18:0). This scheme includes product ions with mass errors <10 ppm.
Discussion
The presence of the described compounds, including FAs and sphingoids, shows that these compounds were either passively transported from the lipid bilayer or cytosol through the mycelial cell walls during the course of the experiment, or actively transported via vesicles (Figure 1). Previous fatty acid profile studies of T. versicolor mycelia mainly include extracts from fruit bodies. Abugri et al. [22] determined the FA composition of wild T. versicolor by hydrolyzing the collected samples with 5 M KOH and MeOH in the presence of the internal standard C19:0. After this procedure, which was performed at 55oC for 90 min, the samples were ice-cooled. The FA methyl esters (FAMEs) that had formed were hexane extracted and analyzed with GC-MS. This study did not investigate any hydroxy FAs, but rather saturated mono-unsaturated (MUFAs) and poly-unsaturated FAs (PUFAs). All of the saturated FAs that were tentatively identified in the present study were also reported by Abugri et al., except for C20:0. The most abundant FAs in the study were C16:0 (palmitic acid), C18:1 (oleic acid) and C18:2 (linoleic acid).
In another study, Rezanka et al. [23] investigated saturated FAs, MUFAs, PUFAs as well as hydroxy FAs in seven species belonging to the phylum Basidiomycota. Among these species was T. versicolor. The fungi were collected in the lake Baikal Ulyanovsk region of Russia. As in the study performed by Abugri et al., C16:0, C18:1, and C18:2 were the most abundant FAs. From the hydroxy FAs, C16:1-OH was identified, but not C18:1-OH and C20:0-OH. These FAs were identified in the present study. Interestingly, in this study, the retention time of C16:1 was shorter than for C16:1-OH (Table 1). It can be explained by a higher degree of branching in C16:1. Rezanka et al. used GC-MS, with two different derivatization protocols. One protocol (including methylation with BF3) was used for non-hydroxyl FAs. Hydroxy FAs were transferred to picolinyl esters -TMS ethers to enable structure elucidations.
Sphingoids form a primary part of the sphingolipids, a class of membrane lipids that include sphingoids, ceramides, cerebrosides, sphingomyelin and gangliosides. The sphingoids contain long- chain aliphatic amines that include two or three hydroxyl groups and often a trans-double bond in position 4 (Figure 3). The sphingoids that are most abundant in fungal ceramides are phytosphingosine and 9-methyl-4, 8-sphingadienine [24]. The [M+H]+ ion of this latter compound, with a molecular formula of C19H37NO2 and a monoisotopic mass of 311.282440 Da, was not detected in the present study. The amines are amide-linked to hydroxylated FAs, which most commonly include 16 or 18 carbon atoms and unsaturation between C-3 and C-4 [25]. The α-hydroxylated C16:0, C18:0 and C18:1 is the three major fatty acyls [26]. The formed amides (ceramides) are linked to cerebrosides by a sugar monomer. Cerebrosides may be present in cell walls as well as in fungal cell membranes, and previous studies have extracted ceramides from fungal cells with chloroform and methanol [27]. In the present study, no ceramides were detected. This may be explained by the mild conditions to which the fungal mycelia were exposed. To the best of our knowledge, this study reports the sphingoid composition of T. versicolor mycelial cells for the first time.
The product ion spectra of sphingoids (sphingosins and phytosphingosins) contain characteristic water losses which determine the number of -OH groups. In the present study, it was experienced that the fragmentation of the back-bone structures included an immonium ion [C2H6N]+. These ions are reported to be present in the fragmentation of amino acids using ESI ionization [28]. It is formed by α-cleavage. In the present study, it was shown that the fragmentation of the sphingoids, which occurred at the amino alcohol moiety, included three mass fragments including 5-7 C separated by on methylene group.
The unsaturated sphingosines Sph (d18:1) and Sph (d16:1) could be distinguished from the saturated ones (Sph d18:0) and Sph d16:0) by the presence of C5H7, C6H9 and C7H11 product ions. The saturated sphingosines contained C5H9, C6H11 and C7H13 product ions. All six sphingoids contained an ion at m/z 55.0542 ([C4H7]+). This ion is often in the literature referred as a “common fragment ion”.
Conclusions
The present study showed that an array of low molecular lipids (<350 Da) leached from the fungal mycelia to the slightly acidic nutrient solution. The underlying reasons can be explained by a passive transport through the plasma membranes or by exocytosis in which vesicle lipids are actively transported to the extracellular environment. The discovered lipids can be considered as harmless to the environment, thereby not preventing the use of the green Trametes versicolor mediated biodegradation procedure.
Acknowledgment
The authors would like to extend thanks to Linda Persson, M.Sc., at GE Healthcare Bio-Sciences AB, Uppsala, Sweden, for her skillful technical assistance.
The authors are also grateful to Ivana Eichlerová, PhD, at the Institute of Microbiology, Academy of Sciences of the Czech Republic in Prague for supplying T. versicolor, and to Elke Hålenius, PhD, at ALS Scandinavia, Danderyd, Sweden for the SEM photographs. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.