- (2009) Volume 10, Issue 4
Alexios S Strimpakos, Kostas N Syrigos, Muhammad Wasif Saif
Yale Cancer Center, Yale University School of Medicine, New Haven, CT, USA
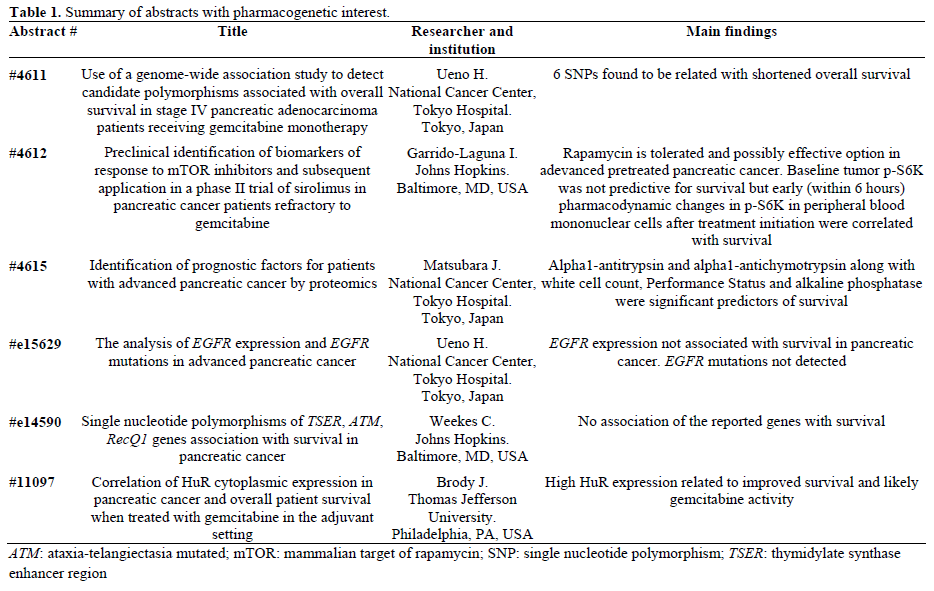
Cancer pharmacogenetics is a popular and evolving field in medicine with applications in various types of tumours helping clinicians to apply a more personalized medicine by providing information of prognostic, predictive and therapeutic value. Such evidence of pharmacogenetic applications is been already available in colon cancer (e.g. KRAS status, mismatch repair genes status, UGT1A1 polymorphisms), lung cancer (EGFR mutations, ERCC1 mutations), breast cancer (HER2/neu overexpression) and many others. In all these tumors, the genetic information is rendering the management of the involved patients safer and more effective. Interesting abstracts and announcements from the perspective of pharmacogenomics in pancreatic cancer included Abstract #4611 which suggested the use of a novel genomic study able to detect specific single nucleotide polymorphisms (SNPs) with prognostic value, Abstract #4615 which showed that the known proteins alpha1-antitrypsin and alpha1-antichymotrypsin may be predictive of response to gemcitabine and survival, and Abstract #11097 which suggested that human R protein (HuR) expression may be a useful predictive biomarker of gemcitabine treatment. The authors also present here a few other abstracts of pharmacogenomic interest which had negative findings, but believed to be of clinical importance
Adenocarcinoma; Pancreatic Neoplasms; Pharmacogenetics
ATM: ataxia-telangiectasia mutated; GWAS: Genome-Wide Association Study; HuR: human R protein; mTOR: mammalian target of rapamycin; SNP: single nucleotide polymorphism; TSER: thymidylate synthase enhancer region
Pancreatic cancer is a relatively uncommon but highly aggressive and lethal malignancy with a very low median survival, which is correctly the perception from the general population [1]. Few risk factors have been implicated in the pathogenesis of pancreatic cancer but it is still unclear what makes this disease quickly progressing. It is known from published literature that many genetic alterations do occur in pancreatic cancer [2]. Among the commonly mutated genes in pancreatic cancer we can find the oncogenes K-ras (75-100%), HER-2/neu (about 65%) p16INK4a (>90%), notch1, Akt-2 and COX-2 but also the tumour suppressor genes p53 (45 to 75%), DPC4 (about 50%), FHIT (70%) and BRCA2. None of these altered genes is currently playing any particular role in clinical practice nor serves as a biomarker of treatment failure or efficacy. In this year’s annual meeting of the American Society of Clinical Oncology (ASCO), few interesting works in the pharmacogenetics field were presented.
Table 1 summarizes the related abstracts and their main findings as reported by the researchers.
“Positive Studies”
The concept and use of Genome-Wide Association Study (GWAS) was presented by Uano et al. (Abstract #4611). The Japanese researchers performed genotypic studies on 67 pancreatic cancer patients stage IV treated with first line gemcitabine monotherapy. DNA analysis was conducted on peripheral blood leukocytes and candidate single nucleotide polymorphisms (SNPs) were detected and associated with overall survival. Of the 11 single nucleotide polymorphisms associated with shorter overall survival, six single nucleotide polymorphisms with a minor allele frequency >5% were found to be statistically significant. These single nucleotide polymorphisms were: rs1861674 in the LOH12CR1 gene, rs10504551 in the TCEB1 gene, rs1132750 and rs2055943 in the GABRA4 gene, rs2589506 in the MAPK10 gene and rs2586404 in the DNM3 gene. The authors concluded that GWAS could be used as a useful tool for identification of biomarkers of survival. Whether the shortened overall survival was related to response to gemcitabine or other reasons was not answered and therefore further prospective studies to confirm these findings are needed.
The same team from Japan presented the results of an interesting study on proteomics as potential biomarkers of response to gemcitabine (Abstract #4615). In this study, the baseline plasma proteome of 29 short-term pancreatic cancer survivors (less than 100 days) was compared with the plasma proteome of 31 long-term survivors (longer than 400 days) by quantitative mass spectrometry technique, from a pool of 304 patients with advanced pancreatic cancer on first line gemcitabine who received at least two cycles of therapy. Out of the 45,277 peptide peaks, 637 were statistically significantly different between the two groups and of those two showed the highest significance, the alpha1-antitrypsin level (odds ratio 3.47; P=0.0003) and the alpha1-antichymotrypsin level (odds ratio 3.00; though P value was reported as 0.160). Other predictors of survival confirmed in this study were performance status, white cell count, platelets count and alkaline phosphatase. The researchers reported that the above prediction model could be used to identify patients who are less likely to benefit from the standard treatment (those with high levels of alpha1-antitrypsin and alpha1- antichymotrypsin) who may need modification of treatment strategy. The above findings have not been reported previously and need repetition by others and validation by larger prospective studies before recommended to clinical practice.
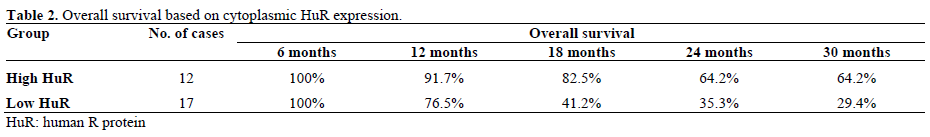
The predictive and prognostic importance of human R protein (HuR) expression was tested in 29 pancreatic patients who received gemcitabine based adjuvant chemotherapy (Abstract #11097). The main actions of HuR protein include RNA stabilization and RNA transportation by binding to mRNA adenine and uracil rich sites (AREs). HuR is a cytoplasmic protein which is upregulated in situations of cellular stress and overexpressed in various solid tumours including aggressive pancreatic adenocarcinoma [3]. This regulating enzyme is also interfering in the metabolism of gemcitabine by regulating deoxycytidine kinase mRNA and therefore overexpression of HuR may increase the activity of gemcitabine [4]. A previous study in colon cancer cell lines has demonstrated that HuR overexpression is associated to high cyclooxygenase (COX) and other angiogenic factors levels [5].
Tissues resected from 29 pancreatic cancer patients were tested for their HuR expression levels. Neoadjuvant chemotherapy was not allowed but adjuvant chemoradiotherapy was given in a few patients. Subsequently, patients were followed up until progression or death. The median overall survival of the studied population was 20.6 months. The median overall survival in the low HuR group was only 15.3 months, whereas median overall survival was not reached yet in the high HuR group at the 40 months follow up point. The hazard ratio of low to high HuR on univariate analysis was 3.36 (P=0.025) going up to 5.04 when adjusted for age, gender and previous radiation therapy. The authors concluded that HuR could provide a useful biomarker of survival and predictor of response to gemcitabine based therapy. It is unknown whether a different adjuvant option excluding gemcitabine could still provide an advantage (e.g. fluoropyrimidine based regimen). In Table 2 we can see the overall survival according to the cytoplasmic HuR expression levels.
“Negative Studies”
In Abstract #4612, the potential use of baseline p-S6K expression (a protein that activates RNA translation initiation in response to stimulation of its upstream TOR regulator) and of early changes of p-S6K in peripheral blood mononuclear cells was tested in patients with advanced pretreated pancreatic cancer treated with temsirolimus, and associated with response to the mammalian target of rapamycin (mTOR) treatment. Though, the baseline levels of p-S6K expression were not associated with survival, the early p-S6K changes were found to be statistically correlated with response to temsirolimus. The clinical use of this knowledge is rather minimal at present as mTOR inhibitors were reported to have no activity in advanced pancreatic cancer in two prospective studies presented in this year’s ASCO meeting (Abstract #4621).
As we know from a recent large phase III study the combination of gemcitabine with erlotinib, an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor offers clinical benefit in terms of overall survival and symptoms improvement. Evidence from lung cancer patients suggests that specific mutations of the EGFR gene (deletion of exon 19 and mutation at exons 18 and 19) predict response to erlotinib. The significance and potential pharmacogenetic application of EGFR expression and EGFR mutations and correlation to survival was tested on tissues from 31 pancreatic cancer patients on treatment with gemcitabine and/or S-1 (Abstract #e15629). Positive EGFR expression levels were detected in 15 patients whereas EGFR expression was negative in 16 patients. In contrast to lung cancer patients, there was no association of EGFR expression to survival (P=0.386) nor evidence of the EGFR mutations seen in lung cancer in the small population tested here. It would be interesting though to confirm these findings in a separate study and to test those in patients treated with erlotinib as well before we dismiss any association of erlotinib activity and efficacy to possible EGFR alterations.
Weekes et al. from John Hopkins Hospital, Baltimore, MD, U.S.A., worked on the hypothesis of association of SNPs of the genes TSER (thymidylate synthase enhancer region), ATM (ataxia-telangiectasia mutated) and RecQ1 to survival and response to chemotherapy (gemcitabine or capecitabine) in advanced pancreatic cancer (Abstract #e14590). The authors also tested whether S/S variant of the TSER gene was specifically associated with higher response to capecitabine. It was reported that no definite conclusion or association of polymorphisms of these genes to response or survival could be drawn, in contrast to previous reports by other researchers few years ago [6]. Nevertheless, the S/S variant of TSER gene was associated with rather higher response to capecitabine but at the expense of excess toxicity which did not justified use of this agent in these patients, according to the authors. The inconsistent results with the previous study on similar setting [6] and the small number of patients are major limitations of this study, thus the results require further evaluation in large cohorts of patients.
Thoughts for Future
Pancreatic cancer remains a challenging solid tumour characterized by an aggressive course, limited therapeutic options and disappointing response to any kind of intervention known so far. In contrast to other tumours, where pharmacogenomic advances have made a difference in clinical practice, by selecting the appropriate population for treatment, the treatment approach in pancreatic cancer patients is rather empirical and guided by positive results of just a few large trials than etiological. Earlier this year, evidence of the predictive value of SNPs of the enzyme cytidine deaminase, involved in the metabolism of gemcitabine, regarding gemcitabine toxicities and the prognostic value of SNPs of mismatch repair genes was presented at the Gastrointestinal Cancers Symposium, 14-17 January 2009, San Francisco, USA. Continuing these efforts, the genome-wide association study presented in this year’s ASCO annual meeting showed that it is possible to identify genes which can predict response to chemotherapy and survival. Major limitations of this study were the small patients’ number it involved, the retrospective nature of the study and the absence of evidence of replication and validation of the results by other researchers. Furthermore, it is not applicable for many cancer units where the majority of cancer patients are looked after. Nevertheless it is a step towards the right direction. On the other hand it seems that EGFR expression is irrelevant to response to treatment or survival and the EGFR mutations seen in lung cancer are not present in pancreatic cancer. Therefore, this common pathway can not be of use in personalized treatment approach yet but of course this observation has to be tested specifically in patients on erlotinib before it is dismissed permanently. Similarly, no clinical value or use as biomarkers can be seen in mTOR pathway molecules since all properly designed studies of mTOR inhibitors have shown negative results.
The clinical utility of alpha1-antitrypsin and alpha1- antichymotrypsin molecules as predictive and prognostic markers need to be confirmed and validated in other studies. If confirmed, these molecules may become quite valuable since the necessary tests for their identification are already in use in many laboratories and more likely to benefit and apply to many cancer patients. Similar validation and repetition of results concern the HuR protein expression and its predictive association with gemcitabine treatment. We have to appraise the great efforts made by researchers and patients in order to improve the outcome of this disease. Clinicians need to join powers with other basic researchers, geneticists, biologists and pharmacists to broaden the knowledge and maximize the results of the research in a stressful and much controlled financially and ethically environment
The authors have no potential conflicts of interest