Introduction
Myeloproliferative disorders (MPDs) are hematological
malignancies characterized by an accumulation of mature cells
in the peripheral blood(1). Usually, Chronic Myeloid Leukemia
(CML), a well characterized entity harbouring the recurrent
t(9;22) translocation and the resulting BCRABL1 fusion gene,
is separated from the classical MPNs such as Polycythemia
Vera (PV), Essential Thrombocytemia (ET) and Primary
Myelofibrosis (PMF), in which a molecular abnormality has
long time been ignored. In the latter group, the recurrent V617F
mutation in the exon 14 of the JAK2 gene has been identified in
2005 and is currently a key marker for MPNs diagnosis as this
mutation is present in 90%, 60% and 50% of PV, TE and PMF
respectively .In 2007, (2)novel recurrent mutations clustered in a
highly conserved region in exon 12 of the JAK2 gene have been
described in patients with PV or Idiopathic erythrocytosis. Exon
14 and exon 12 mutations differ by 2 main characteristics: the
V617F mutation is limited to only one base change (G1849T)
found in all subtypes of MPNs as well as in splanchnic vein
thrombosis and some myelodysplastic syndromes patients
.Exon 12 mutations on the other hands are extremely variable
in sequence and so far restricted to polycythaemia patients.
In a recent study, using allele specific PCR,(3) we reported
the presence of JAK2 exon 12 mutations in 8 out of 24 PV
patients negative for JAK2 V617F mutation, but failed to detect
these mutations in patients with idiopathic erythrocytosis. The
detection of JAK2 exon 12 mutations is technically much more
complicated than V617F mutation detection. Although some
mutants are more frequent than others there has been an increasing
number of different deletions, insertions or base changes described
in the literature since the initial description.(4)
Until recently, mutant detection had to be addressed
either by direct sequencing (of low sensitivity) or allele
specific PCR (of good sensitivity) but inadequate in a routine
diagnosis setting with so many different mutations requiring
multiple individual PCR reactions. High Resolution DNA
Melting curve analysis (HRM) is based on DNA melting in
the presence of saturating DNA binding dyes. Sequence
variants are inferred from changes in the melting
transition of the PCR product as, depending on their
GC content, length or sequence, different PCR products
have different melting temperatures, whether mutated
sequences are known or not. HRM methods have now
been adapted to real-time PCR instruments(5) and,
compared to sequencing or AS-PCR, represent high
throughput and time saving methods with the further
advantage of reducing post-PCR handling of PCR
products. HRM technology has been adapted to the
identification of bacterial species or subtypes, human
SNP genotyping or mutation detection. However,
instruments vary widely in their ability to genotype
variants by whole amplicon melting analysis. Similarly,
several DNA binding dyes may be used with variable
success. Because HRM technology could be a rapid and
convenient tool for detecting the various JAK2 exon12
mutations, we decided to develop one such method with
the prerequisite that it should be reliable enough to give
similar results on 2 different instruments in 2 different
sites. Then, the assay has been further validated on a
cohort of 8 different mutants and 4 non mutated DNA in
2 additional centers, of whom one functioned in a blind
manner.
Patient and methods
Between May 2012 and Dec 2014, suspected MPNs patients
aged 18- 70 years recruited at National center for Cancer and
Research (NCCCR), Hamad Medical Corporation (HMC)
according to WHO 2008 criteria
All procedures were performed in accordance with the
Helsinki Declaration and approved by the Hamad Medical
Corporation research committe.
Peripheral blood and bone marrow samples
3- 5 ml of Peripheral blood(PB) and / or Bone Marrow
marrow (BM) samplesin EDTA were collected every 3 months
and bone samples were collected every 6 months
DNA extraction and quantification
Patients genomic DNA was extracted from whole blood
using Qiagen and Promega DNA extraction procedures
according to the manufacturer's instructions The concentration
and purity of RNA were measured by the nanodrop machine at
optical density (OD) 260/280nmand DNA samples were stored
in -20ºC freezers
Sample Size
The numbers of samples which collected from suspected
MPN’s at 1-36 months of the study are around 3000 samples
Sampling Technique: 3- 5 ml of Peripheral blood in EDTA and
/ or bone marrow samples will be collected from suspected
MPN’s cases Study Subjects.
Recruitment of patients
Suspected 450 MPNs Adult patients 18- 70 years have been
recruited at National center for Cancer and Research (NCCCR),
Hamad Medical Corporation (HMC) according to the following
(inclusion / exclusion) criteria
Polycythemia Vera (PV)
Inclusion criteria 1.Hemoglobin > 18.5 g/dL in men, 16.5
g/dL in women or other evidence of increased red cell volume
2.presence of JAK 2v617f or other clonal abnormalities 3.Serum
erythropoietin: normal or low. 4.Bone marrow biopsy showing
hypercellularity for age with trilineage growth (panmyelosis)
with prominent erythroid, granulocytic, and megakaryocytic
proliferation Exclusion criteria: 5.High eryhropiotein Level.
For Essential thrombocythemia (ET) AND Primary
myelofibrosis WHO 2008 criteria were used as inclusion criteria
Exclusion criteria:
1. not willing to participte in the study
2. not fulfilling WHO criteria for Myelofibrosis
3.age less than 18 or more than 70
JAK2 Mutation screening and quantification
Suspected MPNs cases were screened and quantified for
JAK2 V617F using JAK2 MutaScreenand JAk2Muta Quanta
Kits (Qiagen)
This multiplex assay is based on two probes (double-dye
oligonucleotide labeled with a 5' reporter dye and a downstream,
3' quencher dye with hydrolysis principle). The two probes
designed to detect and quantify JAK J617F and WT.
During PCR, forward and reverse specific primers and two
probes for JAK J617F and WT hybridize and amplify the region
of interest. The hydrolysis probes exploit the 5' →3' exonuclease
activity of the Thermusaquaticus (Taq) DNA polymerase. When
the probes are intact, the proximity of the reporter dye to the
quencher dye results in suppression of the reporter fluorescence
primarily by Förster-type energy transfer. If the target of
interest is present, the probes specifically hybridize /anneal to
its specific target between the forward and reverse primer sites
and is cleaved by the 5'→3' exonuclease activity of the (Taq)
DNA polymerase .The probe fragments are then displaced from
the target, and florescence signal is released and detected. The
increase in fluorescence signal is detected only if the target
sequence is complementary to the probe.
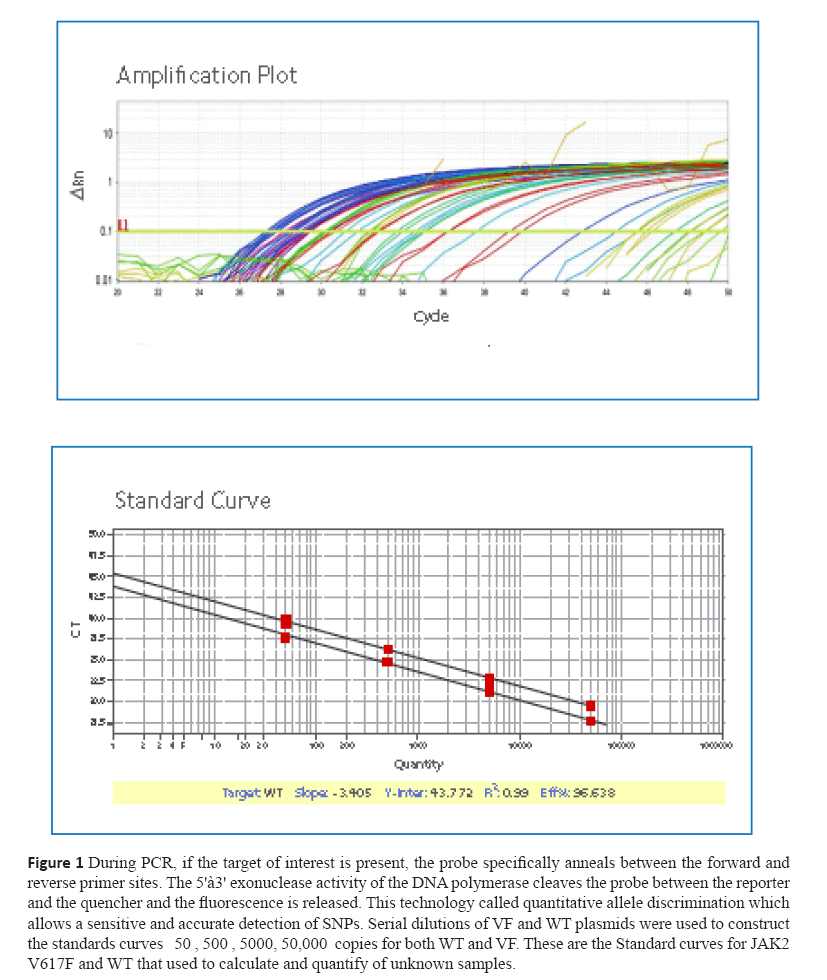
Standard Curves (SCs)
Six serial dilutions (5 x101, 5 x102, 5 x103, 5 x104, copies
/5μls) of JAK J617F and WT and plasmids were used to
establish/construct two standard curves as well as to determine
the concentration of JAK J617F type and WT in patients
samples.
The JAK2 V617F Positive Control (DNA 100% V617F),
V617F Negative Control (DNA 100% WT), and Reference
Sample (DNA 2% V617F) were used.Results interpretation If
the ratio of FAM/VIC for the patient sample is more or equal
to the ratio of reference sample(DNA 2% V617F), the result is
positive and patient has the V617F mutation and if the ratio of
FAM/VICfor the patient samples is less than reference sample
the result is negative and patient doesn’t have JAK2 V617F
mutation
DNA extraction
Patients genomic DNA was extracted from neutrophilis
using QIAamp DNA extraction procedures, and DNA samples
will be stored in -20ºC freezers located in Al- Amal hospitalJAK2
Mutation Screen allelic discrimination assay would be used:
Briefly, Genomic DNA will be extracted from whole blood
or bone marrow and an allelic discrimination assay with two
TaqMan probes will be used (multiplexed assay). One is match
to the allele 1 sequence (eg. the wildtype allele), the other one
is match to the allele 2 (eg. the allele with a mutation). Each
probe is labelled with a distinctive fluorescent dye at its 5’ end
(Reporter) such as FAM or VIC, and contains a nonfluorescent
Quencher at the 3’ end. The probes also contain a minor grove
binder (MGB) permitting the use of shorter probes with greater
stability and thereby a more accurate allelic discrimination.
During the extension phase of the PCR, the perfectly matched
probe is cleaved by the 5’→3’ exonuclease activity of Taq
polymerase, separating the Reporter dye from the Quencher and
thus releasing detectable fluorescence. The mismatched probe
will be displaced rather than cleaved by the Taq Polymerase and
no reporter dye is released. The fluorescence signal (FAM or VIC ) generated is collected at the end of the PCR (endpoint) and
immediately indicates the presence of the targeted sequence(s)
in the sample (wildtype allele, mutated allele or both) in each
experiments. V617F Positive Control (DNA 100% V617F),
V617F Negative Control (DNA 100% WT), and Reference
Sample (DNA 2% V617F) will be used (figure 2) Results
interpretation If the ratio of FAMTM/VIC® for the patient
sample is more or equal to the ratio of reference sample(DNA
2% V617F), the result is positive and patient has the V617F
mutation and if the ratio of FAMTM/VIC® for the patient
samples is less than reference sample the result is negative and
patient doesn’t have V617F mutation
JAK2 exon 12 to 15 mutations screening
The negative MPNs patients for JAK2V617F mutation was
screened to JAK2 exons 12-15
RNA extraction and quantification
RNA was extracted from 1× 107of white blood cells in
guanidiniumthiocyanate buffer (GTC) combined with ionexchange
chromatography using spin-columns (QIAamp
RNA blood Mini kit, Qiagen) according to the manufacturer's
instructions.The concentration and purity of RNA were
measured by the nanodrop machine at optical density (OD)
260/280 nm.
cDNA synthesis:
cDNA was synthesized from 1μg of purified RNA using
reverse transcriptase RT-Dx kit (Ipsogen) according to the
manufacturer's instructions.
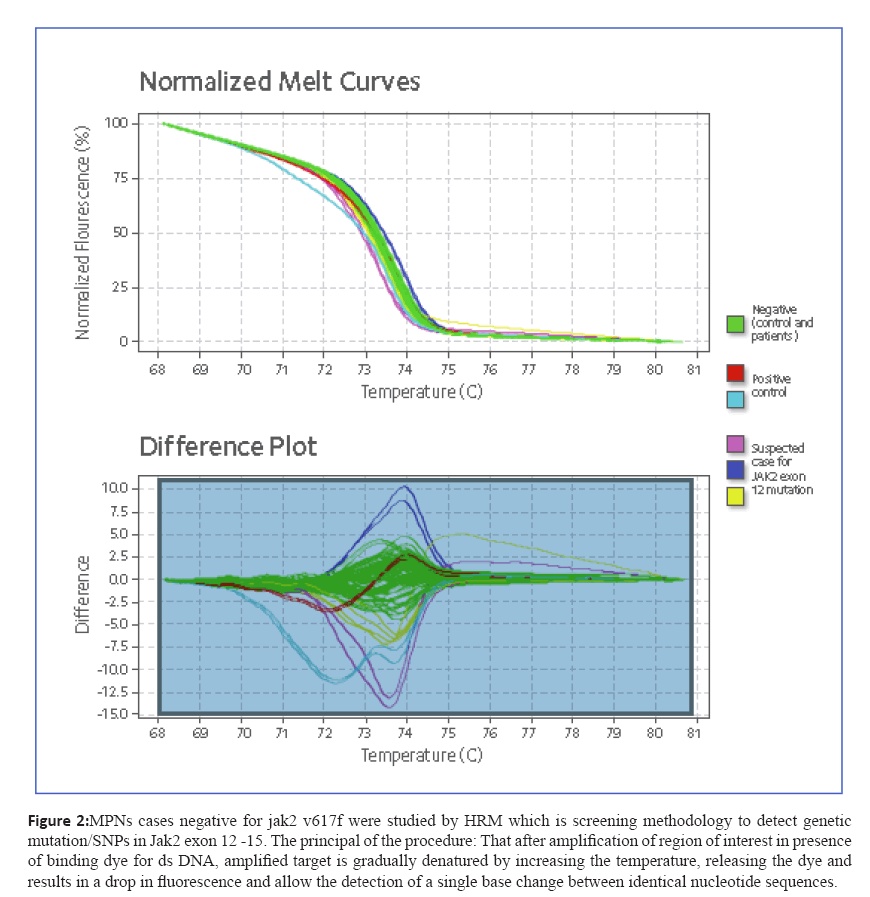
High-Resolution Melting (HRM) Curve Analysis
Amplification reactions and HRM was performed on a
7500 Fast Real-Time PCR System (Applied Biosystems) using
default parameters. Normalized melt curves and difference
plots were analyzed using HRM Software version 2.0 (Applied
Biosystems).
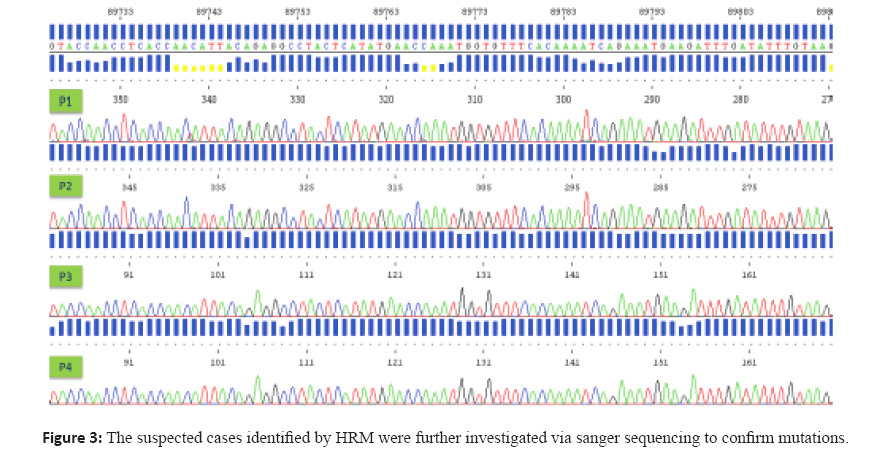
Sanger Sequencing
The suspected cases identified by HRM were further
investigated via sanger sequencing to confirm mutations.
HRM product(8) was treated with ExoSAP or Qiagen PCR
purification Kit, and then the purified PCR product was used
as template for bidirectional sequencing using the BigDye
Terminator Cycle Sequencing Kit (Applied Biosystems,
Carlsbad, CA) on the ABI 3730 Genetic Analyzer. Sequences
are aligned to wild type reference sequence and assessed for
the presence of mutations.
JAK2 (V617F) negative PV patients would be screened
for JAK2 (exon12) mutations using HRM JAK2 exon 12 with
direct sequencing. High resolution melting curve assay Jak2
exon 12 will be amplified and HRM assay will be conducted
to distinguish between the various JAK2 exon 12 mutated
alleles. Sequencing of HRM products HRM product will be
treated with ExoSAPIT or Qiagen PCR purification Kit, and then the purified PCR product will be used as template for
sequencing with the Big Dye Terminator kit.Peripheral blood
and bone marrow will be collected & DNA will be extracted
from suspected ET patients as mentioned on (1.4.1 & 1.4.2)
Suspected ET patients will be screened for JAK2 (V617F)
using RQ – PCR
JAK2 (V617F) mutation as mentioned in section
(1.4.3)1.5.2 JAK2 (V617F) negative ET patients would be
screened for MPL (W515L/K) mutations using RQ-PCR MPL.
MPL Mutations Screen allelic discrimination assay
In an allelic discrimination assay, two different probes will
be used (multiplexed assay). One is a perfect match to the the
wild type allele sequence (eg. the wild type allele), the other one
is a perfect match to the mutation allele sequence Each probe is
labelled with a distinctive fluorescent dye at its 5’ end (Reporter)
such as FAM or VIC, and contains a nonfluorescent Quencher at
the 3’ end. The probes also contain a minor grove binder (MGB)
permitting the use of shorter probes with greater stability and
thereby a more accurate allelic discrimination. During the
extension phase of the PCR, the perfectly matched probe is
cleaved by the 5’-3’ exonuclease activity of Taq polymerase,
separating the Reporter dye from the Quencher and thus
releasing detectable fluorescence. The not matched probe will
be displaced rather than cleaved by the Taq Polymerase and no
reporter dye is released. The fluorescence signal (VIC or FAM)
generated is collected at the end of the PCR (endpoint) and
immediately indicates the presence of the targeted sequence(s)
in the sample (wild type allele, mutated allele or both) (figure
4) 1.5.2.1 Results interpretation: If the ratio of FAMTM/VIC®
patient result is more or equal of ratio of cutoff sample (COS
1.5% of WL/K), result is positive and patient has the WL or
WK mutation and if the ratio of FAMTM/VIC® for the patient
samples is less than result of COS the result is negative and
patient doesn’t have WL or WK mutations
HRM method Three instruments were used in 4 different
French centres: a Light Cycler 480 (Roche Applied Sciences)
in Toulouse and Brest, an ABI 7500 fast (AppliedBiosystems)
in Paris and an ABI 7900 (AppliedBiosystems) in Creteil.
PCR reactions were performed in a 12μl final volume
containing 20 ng of genomic DNA and 0.2μM of forward
(5’- ACCAACCTCACCAACATTACAGAG-3’) and reverse
(5’- AAAAGGACAAAAAAGACAGTAATGAGTATC-3’)
primers defining a 184 bp amplicon. When LightCycler
apparatus was used, the LC480 HRM master mix (Roche),
containing Resolightc as DNA binding dye, was used and 3mM
MgCl2 was added, whereas the AmpliTaq Gold PCR Master
Mix (AppliedBiosystems) with 1.5 μM of Syto-9 (Invitrogen)
were used with the ABI 7500 fast or ABI 7900 instruments.
Amplification was performed by 50 cycles of 95°C for 15
secs, 63°C for 15 secs and 72°C for 25 secs followed by a
melt according to each manufacturer instructions. Sequencing
analysis PCR reactions were performed in a 20 μL reaction
volume containing the following: 10 pmol of each primer forward primer was 5'-CTCCTCTTTGGAGCAATTCA-3' and
reverse primer was 5'- GAGAACTTGGGAGTTGCGATA-3',
1x PCR buffer (Qiagen), 200 μM each dNTPS (Invitrogen), 1U
of HotStarTaq DNA polymerase (Qiagen) and 20ng of DNA.
Cycling conditions were as follows: 95°C for 15 min, 35 cycles
of 94°C for 20 s, 59°C for 20 sec, 72°C for 45s followed by a final
elongation step at 72°C for 10 min. PCR Amplified fragment
was 495 bp in length. Sequencing analyses were performed
using a fluorescent-tagged dideoxy chain termination method
with a 3130XL-DNA sequencing system (Applied Biosystem).
HRM analysis is a suitable method for JAK2 exon 12
mutations detection We previously reported several JAK2
V617F negative polycythemia patients with JAK2 exon
12 mutations detected by allele specific Polymerase Chain
Reaction (AS-PCR(10). In the present study we first used
total blood DNA from these positive patients in order to set
up a new assay using HRM technology in 2 centres. The assay
was first developed on a Lightcycler 480 (Roche) in Toulouse,
and then tested on an ABI 7500 fast (AppliedBiosystems)
instrument in Paris. Using HRM method allowed to identify
every patient known to be mutated, whereas all control
patients showed a wild type profile. These results confirmed
that High Resolution DNA Melting curve analysis is a suitable
method for the detection of JAK2 exon 12 abnormalities. In
order to test for the reproducibility of the method we have
analysed 2 positive samples characterized by different mutant
sequences in 3 independent experiments, each performed
at 1 week interval. Results were highly similar in all three
experiments (not shown), proving that the HRM method
tested herein could have sufficient robustness for a diagnostic
purpose. Though it is now widely admitted that JAK2 V617F
mutations can be detected with similar efficacy in DNA
from peripheral blood or purified granulocytes, this has not
been extensively studied for the JAK2 exon 12 mutations.
We compared in 2 patients the results obtained with DNA
extracted from total blood or from purified granulocytes.
Similar results were observed whatever the source of DNA
Applications of the HRM analysis Samples (n=39) of
patients with either idiopathic erythrocytosis or PV from our
previous report which were negative by AS-PCR for exon 12
mutations were analysed by the HRM method and were again
found negative. In the analysis of new patients addressed
to our laboratories for MPD diagnosis, 9 out of 35 patients
presenting with an increased hematocrit, low Epo levels and
absence of the JAK2 V617F mutation, harboured a mutation
confirmed by sequencing analysis. Validation of the method
in 2 additional centres. It has been reported that important
discrepancies could be observed when one HRM method was
used on different instruments (9). In a preliminary approach,
we thus compared the diagnostic accuracy of our method
by analysing the 9 positive samples on 2 instruments, in 2
separate laboratories in Toulouse and Paris. Each laboratory
used different DNA binding dyes (Resolightc on the LC480
instrument and Syto-9 on the ABI 7500 instrument) also
reported to be source for discrepancies. Although shifting
temperatures and curves shapes were somewhat different
between the two instruments, results were undoubtedly similar in their interpretation. The validation of the method was
completed by analysing 8 samples bearing different mutated
sequences (clinical data and mutant subtypes are given in Table 1) in 2 additional laboratories, Brest and Creteil, which
had not participated in the set up of the HRM assay. In one
of the two labs (Creteil), the analysis was performed in a
completely blind manner, on12 anonymous DNA samples
(8 mutated and 4 wild type) on an ABI 7900 instrument
(Applied Biosystems) whereas a LC480 instrument (Roche)
was used in Brest. The HRM method was equal. (Figure 1),
(Figure 2), (Figure3).
Figure 1: During PCR, if the target of interest is present, the probe specifically anneals between the forward and
reverse primer sites. The 5'à3' exonuclease activity of the DNA polymerase cleaves the probe between the reporter
and the quencher and the fluorescence is released. This technology called quantitative allele discrimination which
allows a sensitive and accurate detection of SNPs. Serial dilutions of VF and WT plasmids were used to construct
the standards curves 50 , 500 , 5000, 50,000 copies for both WT and VF. These are the Standard curves for JAK2
V617F and WT that used to calculate and quantify of unknown samples.
Figure 2: MPNs cases negative for jak2 v617f were studied by HRM which is screening methodology to detect genetic
mutation/SNPs in Jak2 exon 12 -15. The principal of the procedure: That after amplification of region of interest in presence
of binding dye for ds DNA, amplified target is gradually denatured by increasing the temperature, releasing the dye and
results in a drop in fluorescence and allow the detection of a single base change between identical nucleotide sequences.
Figure 3: The suspected cases identified by HRM were further investigated via sanger sequencing to confirm mutations.
Results and Discussion
Results
450 patients were classified into PV, ET & PM. Out of 180
PV, 95% of cases were positive for the JAK2 V617F & 5%
of cases were negative for other mutations. Out of 240 ET,
49% of cases were positive for JAK2 V617F, one had MPL
S505N mutation & 50% of cases were negative for other
mutations. Out of 30 PMF, 35% of cases were positive for
JAK2 V617F & 1 unclassified case was characterized by
DVT had JAK2 exon 13 mutation (R564L). 11 samples were
successfully sequenced, with a mean depth of 1500 reads
& the FASTQC plugin indicated good quality sequencing
metrics. JAK2 V617F, JAK2 exon 12-15 & MPL (S505N,
W515 L/K) negative samples tested before via RQ-PCR,
HRM & sequencing were called negative by NGS. NGS
identified novel deleterious mutations in MPNs patients.
Out of 6 familial cases, 5 patients (P1- P5) were ET & 1
patient (P6) was PV. P1 had JAK2 V617F, ASXL1 T600P,
CBFB G180S, THPO S184R & ITGA2 R76Q, P2 had
JAK2 V617F, MPL A554G & ATM F582L, the other three
Patients (P3, P4 & P5) had CLAR K385fs*47 & one PV
patient (P6) had TYK2 E1163G, ASXL1 P808H, PDGFRB
P4L & TERT G300fs. Among the patients & healthy
individuals, mutations/SNVs such as MPL P106L, K553N,
SH2B3 L476F, ATM F1036F KIT N564S & TET2 T730R
were also found. A complex combination of mutations
in JAK2, THPO, ITGA2 & MPL genes occurred in ET
patients & coexistence of several oncogenic events in
TYK2, ASXL1, PDGFRB & TERT occurred in PV patient.
This finding may also suggest that the MPNs phenotype
may depend on presence of other mutations. It is worth
mentioning that the presence of ATM variant in P2 is
associated with increased risk of CLL. Somatic CALR
type-2 mutation was identified in 3 ET (non mutated
JAK2 or MPL) patients. This mutation is 5-bp TTGTC
insertion in exon 9 that generates a mutant protein with
a novel C-terminal (p.K385fs*47). In patients & healthy
individuals, a heterozygous germ-line mutation in exon 3
of the MPL gene (MPL P106L) has been observed.
Conclusion
Myelooproliferative Neoplasms are prevalent among
Arab Populations with more specific feature of presence
of familial cases as well as predilection to affect younger
age groups which requires further investigations by Whole
exome sequencing or whole genome sequencing to further
explore the disease in this unique category of patients.
Conflict of Interest
This research was conducted as part of the Qatar
national Research Fund-sponsored project “novel approach in Molecular pathophysiology of Myeloproliferative
neoplasms: What determines phenotypes of JaK2 Mutations
(Qatari prospective)” (npRp number 4–471–3–148). this is
applicable to Yassin Ma and al-dewik n.
References
- Passamonti F.Classification of myeloproliferative neoplasms and prognostic factors,AmSocClinOncolEduc Book. 2012:419-24. doi: 10.14694/EdBook_AM.2012.32.419.
- Wadleigh M, TefferiA.Classification and diagnosis of myeloproliferative neoplasms according to the 2008 World Health Organization criteria.Int J Hematol. 2010 Mar;91(2):174-9. doi: 10.1007/s12185-010-0529-5. Epub 2010 Feb 27. Review
- Tefferi A, VardimanJW.Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms.Leukemia. 2008 Jan;22(1):14-22. Epub 2007 Sep 20. Review.
- KuriyamaK.NihonRinsho[Classification of myeloid leukemias].. 2009 Oct;67(10):1853-62. Review. Japanese
- Kouroupi E, Zoi K, Parquet N, Zoi C, Kiladjian JJ, Grigoraki V, Vainchenker W, Lellouche F, Marzac C, Schlageter MH, Dosquet C, Scott LM, Fenaux P, Loukopoulos D, Chomienne C, Cassinat B.Br J Haematol. 2008 Aug;142(4):676-9.
- A Teffari Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithmsLeukemia. 02/2008; 22(1):14-22. DOI:10.1038/sj.leu.2404955
- Francois Girodon, MD, PhD1,Eric Lippert*,2,Sylvie Hermouet, MD, Ph.D.3, andSerge Carillo, PhD High-Resolution Melting (HRM) Curve Analysis Is a Reliable and Relevant Tool InJAK2V617F-Negative Myeloproliferative NeoplasmsNovember 15, 2013; Blood: 122 (21)
- Serge Carillo, Laurent Henry, Eric Lippert, François Girodon, Isabelle Guiraud, Céline Richard, Frédérique Dubois Galopin, Cedric Cleyrat , Eric Jourdan, Robert Kralovics, Sylvie Hermouet, and Thierry Lavabre-Bertrand Nested High-Resolution Melting Curve Analysis A Highly Sensitive, Reliable, and Simple Method for Detection ofJak2Exon 12 Mutations—Clinical Relevance in the Monitoring of Polycythemia J Mol Diagn. May 2011; 13(3): 263–270.
- Zhiyuan Wuequal contributor, Hong Yuanequal contributor, Xinju Zhang, Weiwei Liu, Jinhua Xu, Wei Zhang, Ming Guan: Development and Inter-Laboratory Validation of Unlabeled Probe Melting Curve Analysis for Detection of JAK2 V617F Mutation in Polycythemia Vera PLOS October 20, 2011 DOI: 10.1371/journal.pone.0026534
- Jun Qian, Jiang Lin, Dong-Ming Yao, Qin Chen, Gao-Fei Xiao, Run-Bi Ji, Yun Li, Jing Yang, Zhen Qian Rapid detection of JAK2 V617F mutation using high-resolution melting analysis with LightScanner platform.Clin. Chim. Acta Clin Chim Acta 2010 Dec 19;411(23-24):2097-100. Epub 2010 Aug 19.