Case Series - (2022) Volume 8, Issue 5
Importance of Application of Appropriate Genetic Tests in Day Today Fetal Medicine Clinical Practice: Illustration with Cases
Sreelakshmi Kodandapani1* and
Suryaprakash Rao2
1Malnad Hitech Diagnostic Centre, Shimoga, Karnataka, India
2Lifecell Diagnostics, Chennai, India
*Correspondence:
Sreelakshmi Kodandapani, Malnad Hitech Diagnostic Centre,
Shimoga, Karnataka,
India,
Tel: +91-9686299600,
Email:
Received: 23-Apr-2022, Manuscript No. IPGOCR-22-13113;
Editor assigned: 25-Apr-2022, Pre QC No. P-13113;
Reviewed: 06-May-2022, QC No. Q-13113;
Revised: 10-May-2022, Manuscript No. R-13113;
Published:
16-May-2022, DOI: 10.36648/2471-8165.8.5.21
Abstract
It is important to apply appropriate genetic tests to have the best yield and for complete final diagnosis. Here we
would like to illustrate the importance by demonstrating of cases.
First is a case with major lethal anomalies. Fetus had holoprosencephaly, umbilical cord cyst, and facial dysmorphic
features. After termination, QFPCR was done on fetal tissue. Fetus had trisomy 18: Edward syndrome.
Second case was 12 weeks fetus with cystic hygroma. FISH was done and fetus had turner syndrome. Third was
an interesting one. Fetus had right aortic arch with polyhydramnios. We had dilemma whether to apply MLPA or
CMA. As there was polyhydramnios, we did MLPA first, which was positive for 22q deletion.
The last case was the most interesting one which showed us karyotyping is still the best one and needed for
everyone case was referred for amniocentesis in view of high risk for trisomy 21. Fish report was normal. However,
at karyotyping there was monosomy 12, demonstrating importance of karyotyping and the need for all cases.
Keywords
FISH; QFPCR; MLPA; Karyotyping; Ultrasound
Introduction
Trisomies 21, 18 and 13 are the most common chromosomal
aneuploidy. We usually don’t miss trisomy 18 and 13 by
ultrasound as they have in major malformations of either
heart, kidney and as well as brain in majority of times [1]. But
it is important to confirm diagnosis by genetic testing as this
helps in genetic counseling. Karyotype is the confirmatory
diagnostic test. Although the advantage of Karyotype is, time
tested with 99.9% accuracy, it is time consuming and cell
culture failure is the limitation. Hence rapid aneuploidy tests
are useful. These tests are faster, accurate and also take care of cell culture failure. But it is questionable whether this can
replace karyotyping. Hence we felt to write this write up to
illustrate the importance of each tests.
Case Presentation
Case 1
Fetus at 20 weeks had alobar holoprosencephaly, congenital
diagrammatic hernia and umbilical cord cyst at ultrasound.
Fetal autopsy confirmed these findings. QF PCR was trisomy
18 (Figure 1). Karyotyping was not possible from fetal tissue. Counseling was successful and patient anxiety could be relieved.
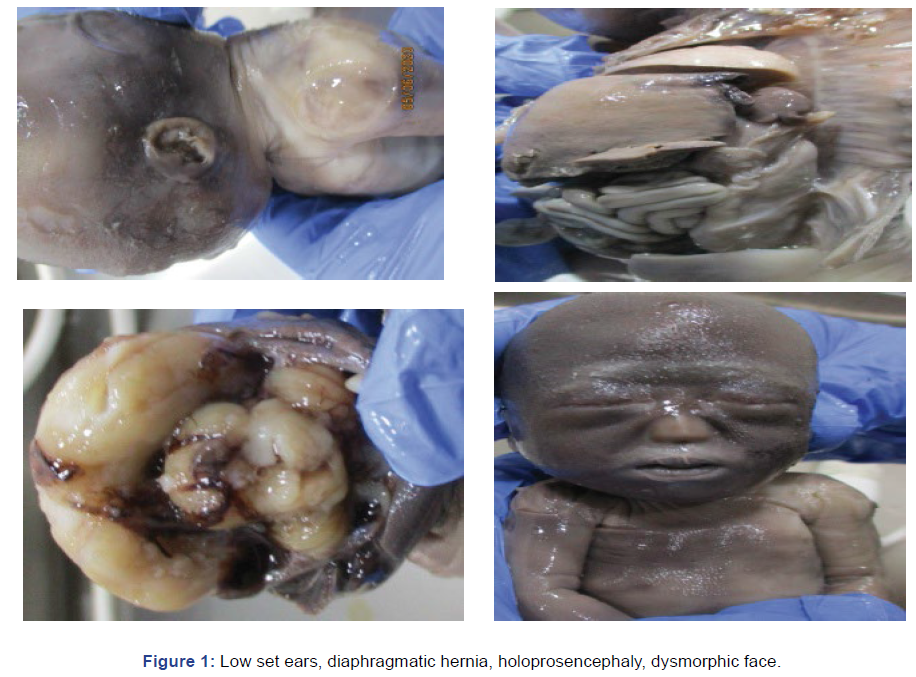
Figure 1: Low set ears, diaphragmatic hernia, holoprosencephaly, dysmorphic face.
Case 2
Fetus at 12 weeks with cystic hygroma was referred or fetal
autopsy and further evaluation. FISH from fetal tissue was
Turner syndrome. Karyotyping was done in this case as FISH
reporting was adequate for fetal evaluation. (Figure 2).
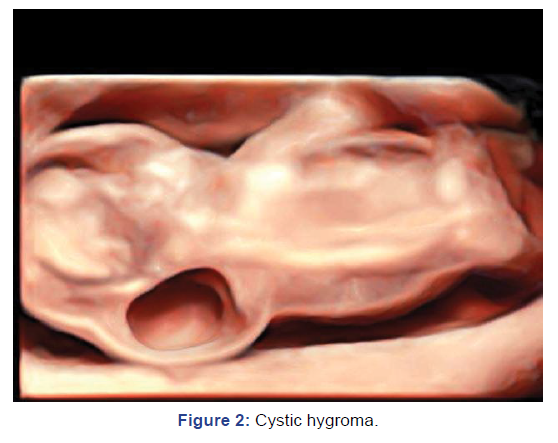
Figure 2: Cystic hygroma.
Case 3
Patient was referred at 20 weeks with right aortic arch and
polyhydramnios. Amniocentesis was done; MLPA and fetal
karyotyping were done. MLPA was 22q deletion (Figure 3).
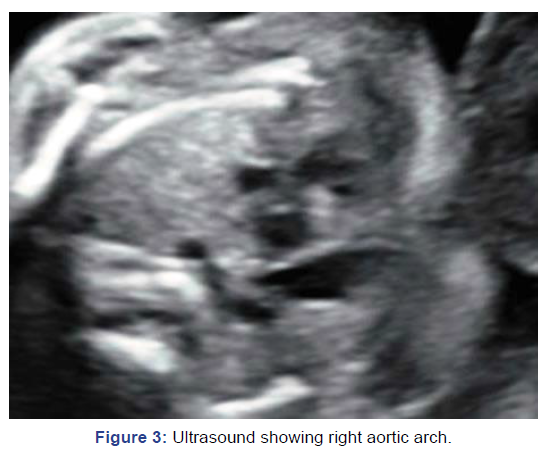
Figure 3: Ultrasound showing right aortic arch.
Case 4
Patient was referred for amniocentesis as there was high risk
for trisomy 21 in Quadruple test. Fetal karyotyping showed
monosomy 12. There were no structural abnormalities in fetus
at ultrasound. At follow up scan there was mild Asymmetric
IUGR, however, prognosis was explained to patient and she is
on regular follow up (Figure 4). It is interesting to know that
mosaic trisomy 12 is rare condition with variable phenotypes.
This is associated with rise in maternal serum AFP and hCG.
There may not be any detectable ultrasound findings. But
many facial features, hearing loss, intestinal malrotation are
described.
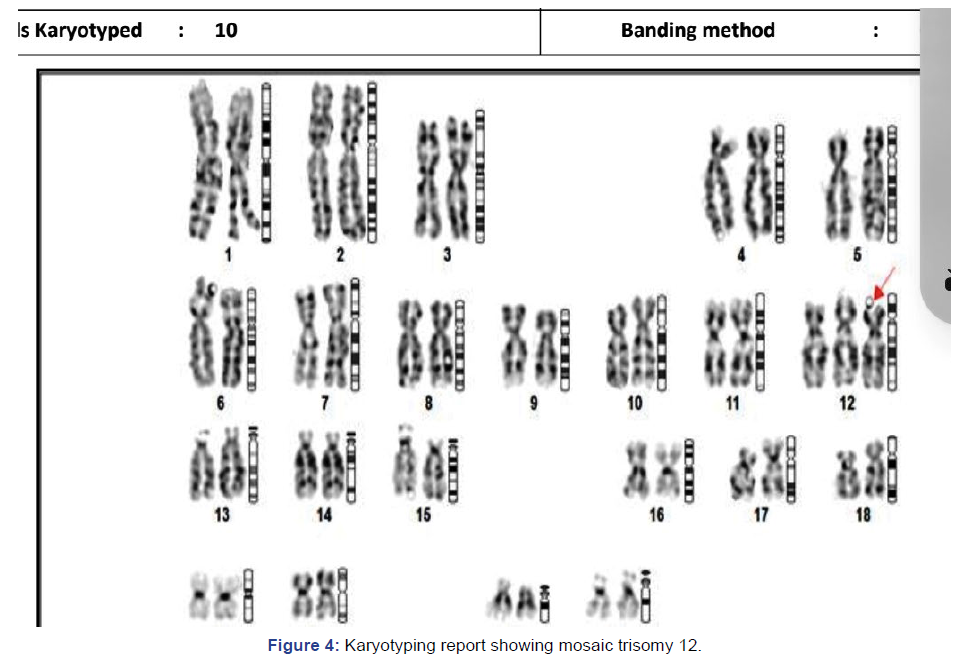
Figure 4: Karyotyping report showing mosaic trisomy 12.
Discussion
It is important to select appropriate genetic testing for correct
result. An ideal genetic test should be highly sensitive, specific,
cost effective and fast. Although a near ideal test is available
and affordable especially in the past ten years, it takes lot of
intelligent input to choose the best appropriate genetic test.
Hence the author felt there is a need to know various genetic
tests, their uses: advantages, disadvantages and limitations
with illustration of cases.
Karyotyping is the gold standard test for nearly half a century.
This is highly reliable test in detection of numeric chromosomal
and major structural abnormalities. These include unbalanced
translocations, balanced translocations (familial and de novo),
mosaicism, supernumerary chromosomes, triploidy and sex
chromosome abnormalities. Karyotyping is a labor intensive
and takes an average of 14 to 21 days. As karyotyping reveals
all chromosomal abnormalities that can be microscopically
detected, it may lead to other findings than the aneuploidy
targeted in prenatal screening. Among those ‘incidental' (or
with a misnomer: ‘unexpected') findings, there can be severe
or mild abnormalities or abnormalities of which the impact on
the health of the child is unsure. Case 4 is the perfect example
for this statement. Patient was referred for amniocentesis, as
there was high risk for trisomy 21 in quadruple test. Karyotyping
was positive for trisomy 12 [2]. Mild fetal growth restriction
was present at follow up ultrasound; however there were no
structural abnormalities. Let us discus clinically relevant points
of each test.
Rapid Aneuploidy Detection Tests
Quantitative Fluorescence PCR (QF-PCR): It is a reliable
molecular method for rapid aneuploidy diagnosis. DNA will be
isolated from the given sample. Multiplex PCR amplification
of Short Tandem Repeat (STR) markers using fluorescently
tagged primer will be carried out [3]. The resulting fragments
will be analyzed on the genetic analyzer for visualization and
quantification. The copy number of each chromosome is
quantified by calculating the relative allele ratio. Analyzed
region includes: D13S252, D13S305, D13S634, D13S800,
D13S628, D18S819, D18S535, D18S978, D18S386, D18S390,
D21S11, D21S1437, D21S1409, D21S1442, D21S1435, and
D21S144. Steps, advantages and limitations are written in
detail in Table 1.
| Genetic Tests |
Steps |
Advantages |
Limitations |
| QFPCR |
DNA extraction
PCR amplification
Capillary electrophoresis
Export data and analysis |
- Large number of samples can be done simultaneously
- Reproducible, easy to perform, and sensitive
|
- Balanced and unbalanced translocations not possible by sensitivity & specificity of the assay may be influenced by the quality of the specimens QF-PCR.
- Samples with significant mosaicism and maternal cell contamination may impact the diagnostic accuracy of QF-PCR
|
| FISH |
Denaturation
Hybridization
Probe detection
Analysis |
- Rapid technique and large numbers of cells can be stored quickly.
- The efficiency of hybridization and detection is high
- Sensitivity and specificity are high
|
- FISH can only detect deletions or duplications of regions targeted explicitly by the probe used and more significant than the probe used. It is possible that FISH may not detect rare, tiny deletions.
|
| MLPA |
Denaturation
Hybridization
Ligation
Amplification
Fragment separation and data analysis |
- Cheap, accurate, known micro deletion and micro duplication can be detected
|
- Cannot detect copy number neutral inversions, translocations, and methylation changes.
- Not a suitable method to detect unknown point mutations.
- Any mismatch in the probe’s target site can theoretically affect the probe’s signal.
|
| Karyotyping |
Culture initiation
Harvest
Hypotonic treatment
Slide preparation, banding and analysis |
|
- Requires fresh tissue
- Although direct preparations can be performed, cell culture is typically required (1–10 days).
- May encounter complex Karyotype with suboptimal morphology.
- Submicroscopic or cryptic rearrangements may result in a false-negative result.
- Normal Karyotype may be observed following therapy-induced tumor necrosis or overgrowth of normal supporting stromal cells.
- Difficulties encountered with bone tumors include low cell density and the release of cells from bone matrix.
- Results are based on the samples received at the laboratory.
- Interpretation is only in correlation with the demographic data provided in the Test Requisition Form.
- Submicroscopic and cryptic chromosome rearrangements cannot be ruled out.
- Partial reproduction of this report is not permitted. However, the contents of this report may be used for research purposes without revealing the personal information of the subject. The excess procedure-related risk above the baseline risk for miscarriage is 0.2 % (1:100)
|
Table 1: Steps, advantages and limitations of genetic tests.
Multiplex Ligation Dependent Probe Amplification (MLPA): Micro deletion and micro duplication syndromes are defined
as a group of clinically recognizable disorders characterized
by a small (<5 Mb) deletion or duplication of a chromosomal
segment spanning multiple disease genes. The phenotype
results from haploinsufficiency for specific genes in the critical
interval. MLPA is used for detection of micro deletion and
duplication.
MLPA is a multiplex reaction, meaning one reaction provides
information on up to sixty targets. For most applications,
a single MLPA reaction is sufficient to answer the specific
questions asked by a physician or researcher. Each MLPA
reaction requires only 50 ng of human DNA. Each MLPA probe
detects a sequence of 60-80 nucleotides, meaning that single
exon deletions and duplications can be seen. Even when MLPA
does not detect any aberrations, the possibility remains that biological changes in that gene or chromosomal region do exist
but remain undetected.
When designing MLPA probes, we exclude any sequences where
known SNVs or other polymorphisms are found using the latest
genetic search engines. This means that a non-pathogenic
polymorphism can also result in a decreased probe signal,
leading to an ambiguous result or, in the worst case, mimicking
a deletion. However, this isn’t always possible, and new
polymorphisms are continuously discovered. We recommend
confirming all MLPA findings with another method, especially
in the case of single probe deletions [4-7].
Fluorescence In-Situ Hybridization (FISH): FISH is a rapid
diagnostic test and detects targeted chromosomes; 18, 13,
21 and sex chromosomes. It is as accurate as QFPCR but more
laborious. However maternal cell contamination can be tackled
with this test [4,8]. FISH test does not provide information
about any chromosome other than the loci mentioned in this
report. A negative result does not exclude the presence of
chromosome alterations other than the one screened for and
imposes the need for a Karyotype. The clinical interpretation
of any test result should be evaluated within the context of the
patient’s medical history and other diagnostic laboratory test
results [9].
Karyotyping: It is the first genetic test and still has a pivotal role
in the genetic diagnosis. The culture failure rate is approximately
3%. In these cases, repeat sampling may be required. Results
may get delayed due to slow cultures. In 3% of cases, placental
mosaicism may be observed. The resolution of the Karyotype
is 5 Mb. Hence genetic abnormalities more minor than 5 Mb
cannot be detected. Normal investigation results do not ensure
a 100% exclusion of genetic abnormalities. Karyotyping relies
on G-band quality and resolution. In general, blood samples
give the best quality chromosomes and, therefore, the best
chance of detecting small subtle chromosome abnormalities.
Chromosomes from other tissues (e.g. amniotic fluid, chorionic
villus and products of conception) give poorer quality
chromosomes; hence the risk of missing a subtle abnormality
increases. It should be noted that on rare occasions, a subtle
abnormality may be missed at prenatal diagnosis, only to be
diagnosed later a postnatal blood sample. The Association of
Clinical Cytgeneticists (ACC) has issued a policy statement to
this effect [10,11].
Caution: It should also be understood that even a G-band blood
Karyotype can never exclude extremely subtle chromosome
abnormalities at the limit of resolution of light microscopy.
Microarray testing should be performed in these cases where
patient meets the appropriate criteria.
The Laboratory adheres to national professional standards for
the minimum acceptable banding resolution for specified types
of clinical referral. If a repeat sample is required due to analysis
failure to meet the minimum standard, the report will state
this.
Although mosaicism may be detected by routine karyotyping, it
can never be 100% excluded. However, if there is an indication
of suspected mosaicism, additional cells will be examined
to exclude 10% mosaicism at a 95% confidence level—
interpretation of mosaicism in prenatal diagnosis.
True mosaicism, when detected prenatally, can be difficult
to interpret, and a further invasive diagnostic test may be
required. Mosaic cell lines may be unevenly distributed
between the fetuses and there is possibility of extra-fetal
tissues leading to false positive and false negative results
in the most extreme cases. Confined placental mosaicism'
(CPM) is observed in approximately 1-2% of CVS samples.
Pseudomosaicism can arise as cultural artifact and does not
represent the fetal Karyotype. This is usually present in only one
or three independently established cultures and can therefore
be interpreted accordingly. In most cases, no further invasive
testing is required. Maternal cell contamination of chorionic
villous and amniotic fluid occurs in approximately 1/250
samples and may occasionally complicate the interpretation of
results.
Normal variation: Each chromosome pair has a specific and
identical G-banding pattern in all individuals. However, variation
of no clinical significance may occur around the centromeric
regions and short arms of some chromosomes. These variations
are known as ‘polymorphic variants’, ‘polymorphisms’ or
‘normal variants’.
Conclusion
RAD techniques are reliable, cheap, fast and cost effective. As
of now these tests don’t need reconfirmation by karyotyping.
However, karyotyping is needed for all prenatal samples,
irrespective of the indication for the invasive procedure. RAD
techniques don’t need reconfirmation from karyotyping and
each other as well.
REFERENCES
- Phadke SR, Gupta A (2010) Comparison of prenatal ultrasound findings and autopsy findings in fetuses terminated after prenatal diagnosis of malformations: an experience of a clinical genetics center. J Clin Ultrasound 38 (5): 244-249.
[Google Scholar], [Crossref], [Indexed at]
- Chen CP, Su YN, Su JW, Chern SR, Chen YT, et al. (2013) Mosaic trisomy 12 at amniocentesis: Prenatal diagnosis and molecular genetic analysis. Taiwan J Obstet Gyne 52 (1): 97-105.
[Google Scholar], [Crossref], [Indexed at]
- Stoiilkovic-Mikic T, Mann K, Docherty Z, Oqilivie CM (2005) Maternal cell contamination of prenatal samples assessed by QF-PCR genotyping. Prenat Diagn 25 (1): 79-83.
[Google Scholar], [Crossref], [Indexed at]
- Schrijver I, Cherny SC, Zehnder JL (2007) Testing for Maternal Cell Contamination in Prenatal Samples. J Mol Diagn 9 (3): 394-400.
[Google Scholar], [Crossref], [Indexed at]
- Rebbeca S, Jo-Ann J, Sylvie L, Wilson RD, Allen V, et al. (2008) New Molecular techniques for prenatal detection of chromosomal aneuploidy. J Obstet Gynaecol Can 30 (7): 617-621.
[Google Scholar], [Crossref], [Indexed at]
- Mann K, Donoghue C, Susan PF, Ducherty Z, Caroline MO (2004) Strategies for the rapid prenatal diagnosis of chromosome aneuploidy. Eur J Hum Genet 12 (11): 907-915.
[Google Scholar], [Crossref], [Indexed at]
- Van Opstal D, Marjan B, Danielle de J, Vandenberg c, Hennie T, et al. (2009) Rapid aneuploidy detection with multiplex ligation-dependent probe amplification: a prospective study of 400 amniotic fluid samples. Eur J Hum Genet 17: 112-121.
[Google Scholar], [Crossref], [Indexed at]
- Tepperberg J, Pettenati MJ, Lese CM, Rita D, Wyandt H, et al. (2001) Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature. Prenat Diagn 21 (4): 293-301.
[Google Scholar], [Crossref], [Indexed at]
- American College of Medical Genetics-Standards and Guidelines for Clinical Genetics Laboratories 2006 Edition
- An International System for Human Cytogenetic Nomenclature (2016). Karger Publishers.
- Human Cytogenetics: Constitutional Analysis: A Practical Approach. 3rd eds. Edited by Denise Rooney.
Citation: Kodandapani S, Rao S (2022) Importance of Application of Appropriate Genetic Tests in Day Today Fetal Medicine Clinical Practice: Illustration with Cases. Gynecol Obstet Case Rep. Vol.8 No.5:21.
Copyright: © 2022 Kodandapani S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.