Short Article - (2017) Volume 3, Issue 3
Qingmei Xie1,2, Shuang Chang1,3, Kunzhe Dong1, John R Dunn1, Jiuzhou Song4 and Huanmin Zhang1*
1USDA, Agriculture Research Service, Avian Disease and Oncology Laboratory, MI 48823, USA
2College of Animal Science, South China Agriculture University, Guangzhou, Guangdong 510642, China
3College of Veterinary Medicine, Shandong Agricultural University, Shandong 271018, China
4University of Maryland, College Park, MD 20742, USA
*Corresponding Author:
Huanmin Zhang
USDA, Agriculture Research Service
Avian Disease and Oncology Laboratory
East Lansing, MI 48823, USA.
Received date: July 18, 2017; Accepted date: August 10, 2017; Published date: August 14, 2017
Citation: Xie Q, Chang S, Dong K, Dunn JR, Song J, et al. (2017) Genomic Variation between Genetic Lines of White Leghorns Differed in Resistance to Marek’s Disease. J Clin Epigenet. 3:29. doi: 10.21767/2472-1158.100063
Genetic resistance to avian tumor virus-induced tumorigenesis and vaccine protective efficacy preventing such tumorigenicity are determined by multiple factors including host genetics, viral virulence, dose of challenge viruses, type of vaccine, vaccine dosage, and interval between vaccination and viral exposure time. Studies on human immune response to vaccination suggest host genetic variability has a strong effect and involves both genes within and outside of the major histocompatibility complex (MHC). Using chickens primarily from two highly inbred and specific pathogen free lines (63 and 72) sharing a common MHC (B*2) haplotype in challenge trials, we recently reported a striking difference in protective efficacy conveyed by HVT or CVI988/Rispens at either 500 PFU/bird or a commercial dosage. We also reported DNA methylation level that differs between the two lines of chickens at promoter regions of genes. Differential gene expression was also reported. This report documents Marek’s disease (MD) incidences of the two highly inbred lines and a series of Recombinant Congenic Strains (RCS) derived from the two lines, which were induced with a very virulent plus strain of MD virus, and illustrates genetic and epigenetic differences between the lines, which we anticipate, at least in part, are liable to the observed MD incidence and vaccine efficacy differences. The genetic and epigenetic mechanisms underlying both genetic resistances to MD and vaccine protective efficacy are complex. Therefore, continuous and systematic efforts on such study are warranted. A better understanding on genetic resistance to MD will empower the disease control through genetic or genomic selection, and a better understanding on the roles of host genetics in relation to immunogenicity in response to vaccination will serve as the touchstone for rational design and development of safer and more efficacious vaccines against infectious diseases.
Keywords
Host genetics; Genetic variation; Marek’s disease; Vaccine efficacy
Abbreviations
ACUC: Animal Care and Use Committee; ADOL: Avian Disease and Oncology Laboratory; BSL-2: Biosafety Level 2; MD: Marek’s Disease; MDV: Marek’s Disease Virus; vv+ MDV: Very Virulent Plus MDV; MHC: Major Histocompatibility Complex; miRNA: microRNA; RCS: Recombinant Congenic Strains; SNP: Single Nucleotide Polymorphism; sRNA: Small RNA
Introduction
Marek’s disease (MD) is a α-herpesvirus-induced T-cell lymphoma in chickens [1,2]. MD historically caused devastating losses to the poultry industry worldwide but has been under control since the 1970s by wide use of vaccination [3-6]. Yet MD continues to cost the poultry industry, with an estimated revenue of over one billion dollars each year worldwide, which are incurred by the routine prevention measures and condemnation, and remains a serious threat to the poultry industry since commercial lines of chickens at large are susceptible to MD and new isolates of MD viruses (MDV) have been emerging with escalated virulence [6-12]. Therefore, current control measures using commercially available vaccines in the same routine as has been practiced during the last half century could be overwhelmed at an unpredicted time point in the future as it happened during the 1980s and 1990s [3,4].
Host genetic resistance is considered as an attractive approach to augment the current control measure against MD [4,13-15]. Host genetic resistance to MD was first noticed in the 1940s [16-18]. It was during and after the 1970s that researchers, using specialized experimental lines of chickens, identified and confirmed that major histocompatibility complex (MHC), the B-haplotypes in chicken, significantly contributes to genetic resistance against MD [19-27]. Soon after, genes outside of the MHC were also shown to contribute to MD resistance [28,29]. Two highly inbred lines of chickens (lines 63 and 72) developed and maintained at the Avian Disease and Oncology Laboratory (ADOL) share a common MHC (B*2) haplotype, but one is highly resistant and the other is highly susceptible to MD as a result of selection [30]. Genetic resistance to MD, either due to the MHC or genes outside of the MHC, is associated with vaccine protective efficacy. Bacon and Witter in the 1990s reported that MHC B haplotypes affect host immune response to MD vaccines. Chickens with B*5 haplotype responded to serotype 2 MD vaccine better and resulted in significantly lower MD incidence than chickens with other B haplotypes. On the other hand, chickens with B*2, B*13, B*15, or B*21 haplotype(s) responded to serotype 1 MD vaccine better than to serotype 2 or 3 MD vaccine [23-26]. In recent years, we have assessed non-MHC genetic variation effect on MD vaccine efficacy using the lines 63 and 72 chickens as well as chickens from a series of recombinant congenic strains (RCS), which all have the same MHC B*2 haplotype [30]. Our data clearly showed genes outside of the MHC region significantly affect MD vaccine protective efficacy. For instance, the two progenitor lines of the RCS, lines 63 and 72, conveyed 72% and 0% protection, respectively, against a very virulent plus strain of MDV (vv+ MDV) challenge in response to HVT vaccination, while the RCS conveyed a range of 43% up to 82% against the same vv+ MDV in response to the same vaccine. Our data also showed for the first time there may be a chicken line by vaccine interaction modulating MD vaccine protective efficacy. We further hypothesized that vaccine protective efficacy, which reportedly depends on many factors, including vaccine dosage, number of vaccinations, age at vaccination, the time interval between vaccination and infection, maternal antibody, type of vaccine, and host genetics, is also partially determined by interactions among vaccine virus genome, host genome, as well as the challenging virus genome [31-33]. Based on the above findings our long-term goal is to determine what are the genetic and epigenetic differences between the genetic lines of chickens that modulate the differential protective efficacy of MD vaccines against vv+ MDV challenge when the other factors listed above are similar or identical. Earlier studies showed the primary lymphoid organ size and IgG level differ between the lines 63 and 72 [34,35]. Data from our recent collaborative studies showed that profiles of global gene expression with or without MDV challenge differ between the highly inbred line 63 and 72 and one recombinant congenic strain [36]. Using miRNA microarray, it was demonstrated that miRNAs were differentially expressed between line 63 and 72 chickens post MDV challenge [37]. DNA methylation level at the promoter region of genes was also found being different between line 63 and 72 chickens with or without MDV challenge [38-42]. The objective of this report is to document the phenotypic differences of MD incidence for the two highly inbred lines 63, 72, and the series of RCS, to probe the genetic and epigenetic variability primarily for the lines 63 and 72, and together to elucidate the genetic and epigenetic variation between the inbred lines that may be responsible, in part, for the phenotypic differences of MD incidence and vaccine protective efficacy at macroscopic level. Our data provided additional experimental evidence in elucidating genetic and epigenetic factors that may modulate MD genetic resistance to MD and the differential immune response to MD vaccine between the genetic lines of chickens.
Materials and Methods
Experimental chickens
Specific pathogen free White Leghorns were sampled from two highly inbred lines (lines 63 and 72) and a series of 19 RCS developed and maintained at ADOL for challenge trials. Chickens from lines 63, 72, and three of the RCS series (RCS-B, RCS-M, and RCS-S) were used as the DNA source for SNP fingerprinting, and chickens from the lines 63 and 72 were used as the RNA source for small RNA sequencing. The lines 63 and 72 carry MHC B*2 haplotype [43]. The RCS were developed from the lines 63 and 72 as previously described [31,40]. Briefly, lines 63 and 72 were crossed followed by two consecutive backcrosses to the donor line 63. Each RCS was then established by sib-mating. Therefore, on average each RCS theoretically possesses 1/8 random line 72 genome and 7/8 line 63 genome [31]. All RCS share the same MHC B*2 haplotype.
Experimental layout
Each challenge trial was conducted by intraabdominally inoculating approximately 30 chicks from each selected line/RCS with a dosage of 500 plaque forming unit (PFU) of 648A passage 40 MDV on the fifth day post hatch. The challenged chicks were housed in negatively pressured BSL-2 isolators. Feed and water were supplied ad libitum. The procedures for sampling and handling of the chickens of the trials were approved by the ADOL Animal Care and Use Committee (ACUC).
Cells and viruses
Primary chicken embryo fibroblast cell cultures were used in preparation of a partially attenuated vv+ MDV, 648A passage 40 [44]. The challenge viruses used in all the challenge trials of this study were from a single batch of preparation.
Postmortem examination
Each challenge trial was terminated 8 weeks post infection. Postmortem examination was conducted for all dead and euthanized chickens to pathologically evaluate for lymphoid organ atrophy, gross tumors and peripheral nerve enlargement. Histopathology exam was employed to confirm diagnoses of MD in all cases of questionable tumors or nerve enlargements.
SNP fingerprinting
Genomic DNA samples were extracted from three chickens per line from the line 63, line 72, RCS-B, RCS-M, and RCS-S. The DNA samples were quantified with a Quant-iT PicoGreen dsDNA Assay (Life Technologies, Grand Island, NY). SNP typing was done with a chicken 60K SNP array [45] at DNA LandMarks Inc. (Quebec, Canada).
Small RNA profiling
Small RNA (sRNA) samples were extracted from thymus tissue of two line 63 and two line 72 chicks at 26-days of age using a miRNeasy Mini Kit (Qiagen, Valencia, CA). sRNAs were ligated to a 5’adaptor and a 3’adaptor sequentially, reverse transcription polymerase chain reaction (RT-PCR) amplified, and used directly for sequencing [46]. The sRNA sequencing was performed on an Illumina HiSeq System (San Diego, CA) by BGI USA Inc. (Woodridge, IL) [47]. sRNA sequences were analyzed following procedures described by Mehta [48] and Shu et al. [49]. Differentially expressed microRNAs were identified using the DESeq package [50].
Results
MD incidence of the line 63, line 72, and 19 RCS chickens induced with a partially attenuated vv+ MDV
The same vv+ MDV, 648A passage 40, induced MD in fewer than 10% of the line 63 chickens but in over 90% up to 100% of the line 72 chickens in two trials (Figure 1). Similar MD incidences for the line 63 and line 72 chickens were also observed in another trial where chickens from the two inbred lines and the 19 RCS were simultaneously challenged with the partially attenuated vv+ MDV. MD incidences for the RCS ranked from 0% (in RCS-J and RCS-L) up to 41% (RCS-M; Figure 2). Since all of the genetic lines 63, 72, and RCS share the same MHC, these MD incidence data clearly demonstrated that genes outside of the MHC complex significantly contribute to genetic resistance to MD.
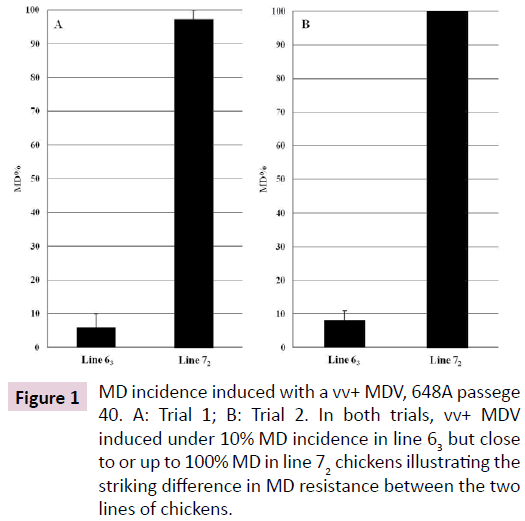
Figure 1: MD incidence induced with a vv+ MDV, 648A passege 40. A: Trial 1; B: Trial 2. In both trials, vv+ MDV induced under 10% MD incidence in line 63 but close to or up to 100% MD in line 72 chickens illustrating the striking difference in MD resistance between the two lines of chickens.
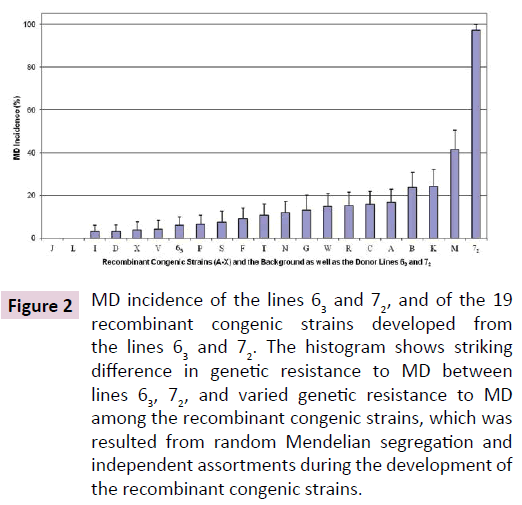
Figure 2: MD incidence of the lines 63 and 72, and of the 19 recombinant congenic strains developed from the lines 63 and 72. The histogram shows striking difference in genetic resistance to MD between lines 63, 72, and varied genetic resistance to MD among the recombinant congenic strains, which was resulted from random Mendelian segregation and independent assortments during the development of the recombinant congenic strains.
SNP fingerprinting of line 63, line 72, and the three RCS
About 98% of the total 57,636 SNPs were typed successfully for all the samples. Of the successfully typed SNPs, 99.9%, 99.5%, 98.6%, 98.6%, and 99.2% were identical by state (IBS) for the lines 63, 72, RCS-B, RCS-M, and RCS-S, respectively. In comparison between lines, about 26.4% of the SNPs in line 72, 6.4%, 4.0%, and 2.8% of the SNPs in RCS-B, RCS-M, and RCS-S differed from line 63, respectively, which reside on chromosomes 1-15, 17- 28 and Z. A Heat map and Dendrogram depicted the relative distance among the lines. The line 63 is clearly most distant from the line 72, followed by RCS-B, RCS-M, and RCS-S (Figure 3). The SNP fingerprint results clearly demonstrated the very existence of genetic variation at the DNA level between the progenitor lines, and even between the progenitor background line 63 and the RCS.
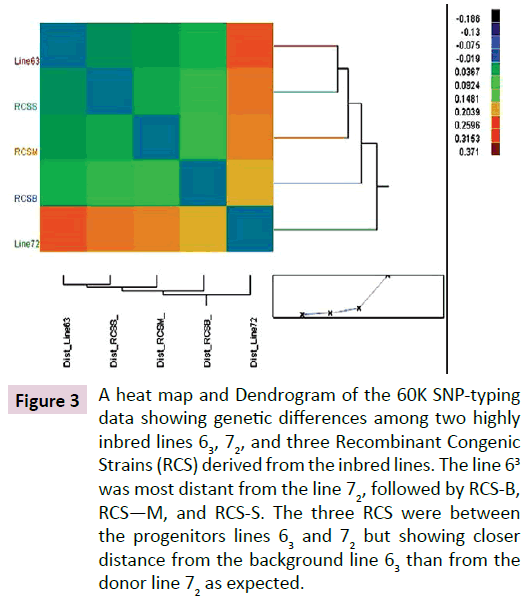
Figure 3: A heat map and Dendrogram of the 60K SNP-typing data showing genetic differences among two highly inbred lines 63, 72, and three Recombinant Congenic Strains (RCS) derived from the inbred lines. The line 63 was most distant from the line 72, followed by RCS-B, RCS—M, and RCS-S. The three RCS were between the progenitors lines 63 and 72 but showing closer distance from the background line 63 than from the donor line 72 as expected.
Preliminary profiling of sRNA expression in thymus tissues of line 63 and 72 chicks at 26 days of age
Deep sequencing identified over a million unique sRNA for each of the chicks from both lines 63 and 72 when all reads were aligned against databases including mirBase (Release 18, Nov. 2011), rRMA etc. (Rfam10.1, NCBI GenBank), mRNA, Target, Repeat (https://hgdownload.cse.ucsc.edu/goldenPath/galGal3/ bigZips/chromOut.tar.gz), and Genome (https://hgdownload. cse.ucsc.edu/goldenPath/galGal3/bigZips/chromFa.tar.gz). Only about 11.3% of sRNAs were found in common between line 63 chicks and 13.9% of sRNAs between line 72 chicks (Figure 4). High percentages of sRNAs differed between line 63 and line 72 chicks, which ranged from 42% versus 45% (line 63 bird 1 versus line 72 bird 2) to 33% versus 55% (line 63 bird 1 versus line 72 bird 1) (Figure 5). The preliminary analysis of the sRNA data indicated that small non-coding RNAs differ in expression in the chickens within and between the highly inbred lines 63 and 72.
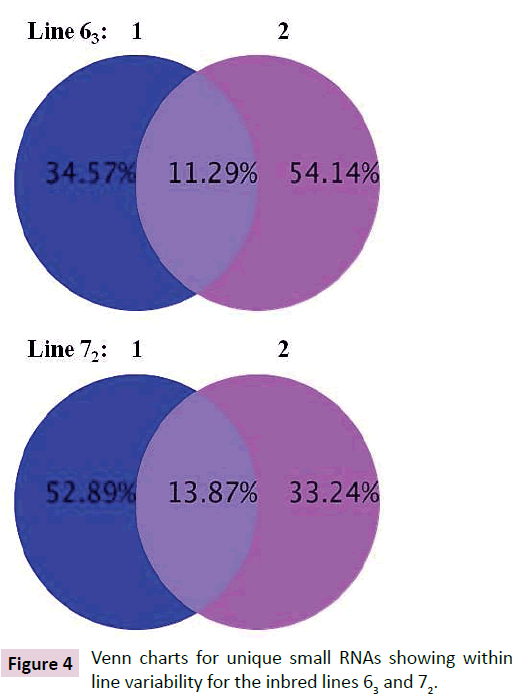
Figure 4: Venn charts for unique small RNAs showing within line variability for the inbred lines 63 and 72.
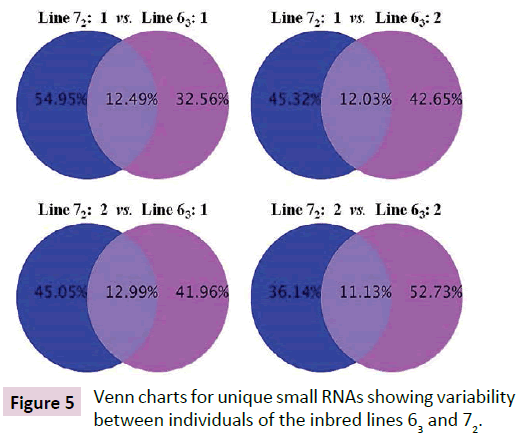
Figure 5: Venn charts for unique small RNAs showing variability between individuals of the inbred lines 63 and 72.
Discussion
It is common knowledge to the poultry research community and the industry that vaccination is of vital importance to protect poultry against MD that once devastated and still remains a serious threat to the industry worldwide [3]. It is also hard to imagine that the poultry industry could have enjoyed the success and prosperity as it has been during the last 40 years without the invention and proper implementation of varied vaccines to efficiently control a variety of infectious diseases of poultry [51,52]. By the same token, it holds pretty much true, if not more true, in human medicine as well [53]. Yet the challenge for maintaining such a good level of control and even tougher a challenge for improving the control against infectious diseases remains. Many factors affect genetic resistance to MD and vaccine protective efficacy. Some of these have been well investigated and reported [23-26,54-56]. We recently showed non-MHC genetic variation, in addition to MHC B haplotype that was primarily reported by Bacon and Witter in the 1990s [23-26,57], also plays an important role in modulating MD vaccine efficacy [31-33]. The findings of MHC and non-MHC effects on MD vaccine protective efficacy are in good agreement with reports of studies both in humans and other animals [58,59]. Line 63 is resistant to MD while line 72 is highly susceptible to MD. Figures 1 and 2 documented the striking difference in MD incidence between the two genetic lines of chickens challenged with a partially attenuated vv+ MDV, 648A passage 40, in three consecutive trials. The striking difference was also observed in HVT protective efficacy between the two lines of chickens. Under a commercial dosage, two HVT vaccines (from two different manufacturers) conveyed protection well over 80% for line 63 chickens challenged with low passage vv+ MDV while the same two HVT vaccines provided 0% and10% protection, respectively, in line 72 chickens under the same challenge [32]. Evidently, host genetic resistance to MD is highly and positively associated with HVT protective efficacy. Furthermore, genetic and epigenetic differences that exist between the two inbred lines of chickens may underlie not only genetic resistance to MD but also host genetics effect on vaccine protective efficacy. The exact genetic and epigenetic factors that confer genetic resistance and/or vaccine efficacy remain to be investigated in addition to what have been reported [60]. SNP-fingerprinting showed that line 63 differs at the DNA level from line 72. The same SNP-fingerprint showed three RCS (RCS-B, RCS-M, and RCS-S) were more different from line 72 than line 63 as expected (Figure 3). Our recent study on gene expression profiling indicated that global gene expressions were significantly different between the chickens of lines 63 and 72 as well [36]. In addition to genetic differences, epigenetic differences now have been found to commonly exist between the two lines of chickens. A significant portion of genes that have been assessed differ in DNA methylation level at CpG sites within the CpG islands of promoter regions between the line 63 and 72 chickens [38-42]. Difference in microRNA expression was also detected using chicken microRNA array between the two lines of chickens [37]. The results of preliminary analyses of small RNA data acquired from Illumina’s HiSeq deep sequencing in this study, which include microRNAs, suggested there is abundant variability within as well as between line 63 and 72 chickens (Figures 4 and 5). This finding is in good agreement with our earlier report [37]. The differences in epigenetic factors, DNA methylation, histone modification [61], and small RNA expression potentiated the possibility that the underlying mechanism conferring resistance to MD and host effect on vaccine protective efficacy may be pretty complex and potentially involving all or a different combination of the factors that differ between the two genetic lines. This may very well hold true, more or less, in human medicine with regard to disease resistance and immune response to vaccination at the personal medicine level.
In summary, we propose that host genetic resistance to MD and host genetics effect on vaccine protective efficacy are highly likely determined by genetic and epigenetic factors jointly. The genetic factors include genomic DNA polymorphisms (SNPs, deletions, and insertions, translocations) and gene expression; epigenetic factors include DNA methylation, histone modification, and non-coding (long and small) RNA expression. Both genetic and epigenetic factors likely encompass MHC and non-MHC genomic regions. The line 63 and 72 chickens are an important genetic resource for poultry research, which should be instrumental not only for identifying genes, pathways, and specific epigenetic factors underlying genetic resistance to MD, but also to host genetics effect on vaccine protective efficacy. Identification of specific genes, pathways, and epigenetic factors affecting genetic resistance and vaccine protective efficacy is of great importance for sustaining current control of the infectious disease and for development of new and more efficacious vaccine, which could continue to empower the poultry industry to maintain good control of the disease and to provide quality chicken products for consumers.
Acknowledgements
This work was partially supported by an USDA-NRICGP grant 2008-35204-04660 (awarded to J.S. & H.Z.) and by funds from National Research Sponsored Project-8 (NRSP-8).