Research Article - (2025) Volume 15, Issue 1
Received: 05-Oct-2023, Manuscript No. EJEBAU-23-17883; Editor assigned: 09-Oct-2023, Pre QC No. EJEBAU-23-17883 (PQ); Reviewed: 24-Oct-2023, QC No. EJEBAU-23-17883; Revised: 14-Jan-2025, Manuscript No. EJEBAU-23-17883 (R); Published: 21-Jan-2025, DOI: 10.36648/2248-9215.15.1.39
Bio-surfactant and red pigment producing, a special bacterial strain was isolated from oil containing soil sediment sample, collected from the Ennore Coastal Region which is located beside the Coromandel Coast of the Bay of Bengal. The potential strain was identified as Serratia marcescens MT053078 based on the biochemical and physiological tests together with 16S rRNA sequence analysis. Maximum production and culture conditions were optimized with the support of emulsification index test through different parameters such as pH, temperature, rpm, salinity, carbon/ nitrogen sources, metal ions and different edible oils. Extraction, purification and characterization of bio-surfactant succeeded by medium fermentation process in optimized condition. Thin Layer Chromatography (TLC) and Fourier Transform Infrared Spectroscopy (FT-IR) were employed to confirm the class of bio-surfactant as lipopeptide. A peak of absorption at about 265 nm was detected in the UV-visible spectrum. The emulsification index activity of Cell Free Supernatant (CFS) with kerosene oil was found to be above 80% by the recommended formula. The results proved that the extracted biosurfactant has potential efficacy against clinical pathogens, as evidenced by the antibacterial and antibiofilm activities.
Serratia marcescens MT053078; Bio-surfactant; Antibacterial; Biological evaluation
The traditionally using synthetic surfactants are highly toxic to aquatic system, because there is insufficient rate in biodegradation. However they are extremely valuable and important chemical products which are utilized in large amount worldwide for various purposes [1]. Surfactant molecules are amphiphilic in nature consisting of a hydrophobic tail and a polar head. In the preparation of medicines, cosmetics and detergents surfactants are used as an active ingredient. Due to this context scientific attention and challenges has been highly increased presently on the extraction of bio-surfactant from microbial resources [2].
A surface active molecule produced by microbes (bacteria, fungi and virus) are called as Bio-Surfactant (BS), they are heterogeneous group of secondary metabolites. In the nature BS occurs as chemical entities such as fatty acids, glycolipids, lipopeptides/lipoproteins, neutral lipids, phospholipids, polymeric and particulate surfactants [3]. Bio-surfactant gained many considerable scientific attentions by their biodegradability, low toxicity, higher salinity, emulsion forming/breaking capacity [4]. Bio-surfactant also has emerged as a prospective agent in food processing and health care industries. They are also playing a vital role in the fields of biomedical, pharmaceutical, cosmetic and agriculture. Due to their high stability to different parameters like pH, temperature and salinity, BS is not only used as antifungal, anti-bacterial and antiviral compounds, they also used as anti adhesive, anticancer and anti-biofilm agents. The Gram negative and motile bacterium S. marcescens is a facultative anaerobe; it can survive in both aerobic and anaerobic conditions. In this study the extraction and purification of bio surfactant, synthesized by S. marcescens MT053078, using liquid paraffin as substrate was investigated [5].
Collection of Sample
Crude oil contaminated sediment sample was collected using sterile spatula at the depth of 5 to 10 cm earth surface from the shores of Ennore creek and coastal region. Then the sterile polyethylene bag containing sample was aseptically transported to the laboratory and stored at suitable temperature in the refrigerator before they are used for the isolation of bio surfactant producing bacteria [6]. Isolation of bacterial cultures carried out in general instrumentation laboratory at Loyola Institute of Frontier Energy, Loyola College, Chennai.
Isolation of Bacteria
Serial dilution technique was adopted to isolate the individual colonies. One gram of sample was mixed with 10 ml of sterile distilled water and the mixture was further diluted serially up to six folds from 10-1 to 10-6. Then 0.1 ml (100 µl) of aliquot from 10-6 was inoculated on to the surface of sterile nutrient agar medium which was already got solidified in petri plates and a sterile L-shaped glass rod was used to spread over the aliquot evenly on agar medium [7]. The plates were then sealed with plastic wrap and incubated at 37°C for 24 hrs. After the desired incubation period, morphologically different individual colonies which were grown on the medium were sub-cultured. The cultures were maintained on nutrient agar slants in test tubes containing (g/L) peptone 5.0, beef extract 3.0, NaCl 5.0 and agar-agar 20.0. Final pH of the nutrient slant was set to be 7.2 and they were stored in refrigerator as a stock culture for further tests.
Screening for Bio-Surfactant Producers
There were totally ten different individual cultures including one red-pigment producing bacteria isolated. All those isolates were subjected to the following five different screening methods and the bio-surfactant producing efficiency of the isolates was evaluated [8].
Blue-agar plate assay: The blue agar medium supplemented with glucose (2%) as a carbon source, cationic surfactant Cetyl Trimethyl Ammonium Bromide (CTAB-0.5 mg/ml), basic dye methylene blue (MB-0.2 mg/ml) and the solidifying agent agar-agar (20 g/L) was used to identify the anionic bio surfactant producer. A loop full of bacterial culture was streaked on the blue agar medium. Then the plates were incubated at 37°C for 48-72 hrs. After the desired incubation period, formation of dark blue zone around the culture was considered as positive result for the production of anionic bio surfactant.
Drop collapsing technique: In the drop collapsing assay, 2 µl of liquid paraffin was added inside the lid of 96-well micro titre plate. The plate was equilibrated for 24 hrs at 37°C and the next day 5 µl of the cell free supernatant centrifuged from the pre grown broth was added to the surface of the paraffin oil. After 1 min, shape of the drop added on the surface was observed. This method was examined using distilled water as a control, the drop which got collapsed by cell free supernatant was denoted as a positive result, whereas the drop remain beaded was noted as negative.
Emulsification index: The emulsion producing ability of bio surfactant with some liquid hydrocarbons was determined by using kerosene. The bacterial culture was inoculated in the nutrient broth and incubated for 24 hrs at 37°C. The cell free supernatant was obtained by centrifugation at 6000 x g at 4°C for 20 min. One ml of kerosene mixed with 1 ml of supernatant and vortexes with high speed for 2 min. Then the mixture was allowed to stand for 24 hrs. On the next day the emulsification index was calculated (Formula 1) by total height of the liquid divided by emulsion height and multiplying by 100.
E24 (%)=Total height of the solution/ Total height of the emulsion × 100 (1)
Hemolytic activity: Blood agar medium was prepared with the composition of casein enzymic hydrolysate (14 g/L), peptone (4.5 g/L), yeast/beef extract (4.5 g/L), NaCl (5 g/L) and agar-agar (20 g/L). After autoclave, sheep blood (5 ml/L) collected (anticoagulant EDTA added) from the slaughter was mixed with the warm medium and poured in petriplate. A loop full of bacterial culture was streaked on the blood agar medium and incubated for 48-72 hrs at 37°C. Results recorded based on the type of zone observed i.e., α hemolysis: The colony surrounded by green color zone, β hemolysis: Zone of clearance around the culture and γ hemolysis: Absence of any change around the colony.
Lipase enzymatic assay: The isolated bacterial cultures that produce lipase enzyme were selected using tri-butyrin agar plates. The medium was prepared with 1% tributyrin and pH of the medium was adjusted to 7.3-7.5 using 0.1 N NaOH. A loop full of inoculum was inoculated on the tri-butyrin agar plates. They were then incubated at 37°C for three days. After incubation period, plates were observed for the formation of clear zone around the culture to select the bio-surfactant producer.
Oil displacement method: Twenty four hours grown inoculum in nutrient broth was used for this screening method. The base of petriplate was filled with 40 ml of distilled H2O. On this water, 10 µl of liquid paraffin was placed as a tiny bead. Further 10 µl of the cell free supernatant obtained by centrifugation with high speed, was added on the oil bead which was located on the distilled water. After few seconds the occurrence of clear zone with blasting the oil bead was indicated as a positive result for bio-surfactant producer.
Antibiotic Sensitivity and Phenotypic Characterization
A loop full of bacterial culture was inoculated in 50 ml nutrient broth and incubated for 6 hrs at 37°C. After the desired incubation period 1 ml of culture broth was taken in a test tube and turbidity of the same was matched with McFarland standard (0.5%) by using sterile water. From this 100 µl of broth was inoculated on nutrient agar medium and the sample was spread over the medium with the help of L shaped glass rod [9]. After few minutes different antibiotic susceptibility discs (HiMedia Laboratories, Mumbai, India) were placed on the swab with the help of forceps. The plate was incubated at 37°C for 24 hrs, on the next day the zone of clearance was measured by zone scale. To identify the characteristics of isolated strain, motility test, gram staining and some other biochemical tests were done (Table 1).
|
Phenotype/Genotype |
Character |
|
16S rRNA (GenBank Acc. No.) |
MT053078 |
|
Color |
Red |
|
Gram character |
- |
|
Motility |
Motile |
|
Shape (compound microscope) |
Rod |
|
Utilization of sugars: Sucrose, glucose |
+ |
|
Gelatin hydrolysis |
+ |
|
Deoxyribonuclease (DNase) |
+ |
|
Lipase |
+ |
|
Catalase |
+ |
|
Pigment (prodigiosin) |
+ |
|
Note: (-) indicates negative and (+) indicates positive |
|
Table 1: Morphological, biochemical and genetic characteristics of Serratia marcescens MT053078.
Genotypic Characterization of Serratia marcescens MT053078 by 16S rRNA Analysis
Genomic DNA was isolated using NucleoSpin® tissue kit (Macherey-Nagel) and the following chemicals and materials were used. A small part of bacterial culture, micro centrifuge tube, 180 µl of T1 buffer, 25 µl of proteinase K, 5 µl of RNase A (100 g/ml), 200 µl of B3 buffer, 210 µl of 100% ethanol, NucleoSpin® Tissue column, 2 ml collection, tube, centrifuge (11000x g), 500 µl of BW buffer, 600 µl of B5 buffer, 1.5 ml tube, 50 µl BE buffer for DNA elution)
Agarose gel electrophoresis was used to check the quality of eluted DNA with the help of following materials and reagents such as 1 µl of 6X gel-loading buffer (0.25% bromophenol blue, 30% sucrose in TE buffer pH-8.0), 5 µl of DNA, 0.8% agarose gel, 0.5X TBE (Tris-Borate-EDTA), 0.5 µg/ml ethidium bromide, 0.5X TBE (at 75 V), UV transilluminator (Genei), UV Gel documentation system (Bio-Rad). Then the Polymerase Chain Reaction (PCR) was carried out in a PCR thermal cycler (GeneAmp PCR System 9700, Applied Biosystems). The following substances were involved in PCR for 35 cycles such as 2X phire master mix (5 μL), D/W (4 μL), forward primer (0.25 μL), reverse primer (0.25 μL) and DNA (1 μL). The PCR product was subjected to agarose gel electrophoresis and sequence analysis, all these steps were done in Rajiv Gandhi Centre for Biotechnology (RGCB), Kerala, India.
Culture Conditions Optimization
The production of secondary metabolites and cell growth were strongly influenced by making mild changes in the composition of medium such as sucrose, nitrogen, carbon, temperature and pH. The optimization of these factors can produce more metabolites and the process was done by one factor at a time [10]. The maximum production was observed by emulsification index. All the reagents and media used for this study were obtained from HiMedia, Mumbai, India.
pH: This is one of the most important characteristics of microorganism. The production of secondary metabolites and cell growth can be improved by giving some changes to the pH of culture medium. The pH values were adjusted from pH 4 to pH 9.5. The bio-surfactant producing efficacy was measured by emulsification index.
Temperature: One of the critical parameter in medium optimization is temperature. Bacterial culture was grown in the temperatures ranging from 20°C to 65°C. After the desired incubation period the emulsification index was calculated.
Agitation: Though the strain S. marcescens MT053078 is a facultative anaerobic organism, the effect of agitation was carried out to check the efficiency of bacteria by using orbital shaker. The rpm speed was adjusted from 100 to 200. In another flask a bacterial culture was inoculated and incubated at static condition. The emulsification index was calculated as mentioned earlier.
NaCl: The assessment of salt tolerance limit of microbes is an important task. The salinity of the broth was adjusted from (0.2% to 3%). The best salt tolerance limit of bacteria to produce maximum bio-surfactant was measured by emulsification index.
Carbon source: Different types of carbon sources such as fructose, glucose, lactose, sucrose and tryptone were added with 0.1% of liquid paraffin and bacterial culture was inoculated. After incubation period a best carbon source was selected by emulsification index which is described as earlier.
Nitrogen source: In the process of bio-surfactant production by microbes, nitrogen plays a vital role. Different varieties of nitrogen sources such as beef extract, casein enzymic hydrolysate, peptone, sodium nitrate and yeast extract were used in the broth to select one of the best among them. Emulsification index was measured after the incubation.
Metal ions and oils: Some important metal ions and edible oils were added with the medium to enhance the growth bacteria and the production of targeted secondary metabolite bio-surfactant. Simultaneously emulsification index test was employed to analyse the bio-surfactant producing activity.
Mass cultivation: The bacterial cells ware grown in 1000 ml (1 L) Erlenmeyer flask with the presence of optimized compositions. After 24 hrs of incubation the cells were removed by centrifugation (6000 rpm x g) at 4°C for 30 min. The pellet was discarded, CFS collected and the pH was acidified (pH-2) with 5N HCl [11]. Then the sample was incubated in refrigerator at 4°C overnight. On the next day precipitate was observed and they were collected by centrifugation.
Extraction of Bio-Surfactant
The crude bio-surfactant was extracted thrice with (100 ml) methanol by shaking the mixture vigorously with the help of separating funnel. When methanol was added as a solvent the extract was immediately neutralized to avoid the formation of methyl esters [12]. The organic layer which contains the precipitated bio-surfactant was allowed to evaporate. Crude bio-surfactant secreted by S. marcescens MT053078 was extracted from post 24 and 48 hrs of incubation.
Partial Purification and Lyophilization
The crude biosurfactant was dissolved in 0.05 M sodium bicarbonate. After filtration, the pH of this solution was adjusted to pH-2 using 5 M HCL and then the solution was kept at 4°C to 8°C for 24 hrs. The precipitate was finally collected by centrifugation at 6,000 rpm for 15 min and lyophilized [13]. The culture is rapidly frozen at 70°C and then dehydrated by vacuum and the tubes containing freeze dried product are stored in refrigerator at 4°C for further use.
Characterization of Purified Bio-Surfactant
TLC (Thin Layer Chromatography): Silica gel G coated commercially available plate was used and the partially purified BS was taken as a stationary phase for TLC. The following solvent composition was poured in TLC chamber as a mobile phase (Chloroform: Methanol: Water) with the ratio of 75:15:5. Few drops of crude BS was loaded over the silica plate with the help of capillary tube and then mobile phase was allowed to run. The separated spot was visualized by UV chamber and scraped out for further characterization [14]. Meanwhile, ninhydrin reagent (100 ml) was prepared to visualize the lipopeptide bio-surfactant by spraying them over another plate.
FTIR analysis (Fourier-Transform Infrared Spectroscopy): The purified bio-surfactant was analyzed in Shimadzu IRTracer-100 FTIR-spectrophotometer, in Attenuated Total Reflectance (ATR) mode considering a range of 500 to 4000 cm-1 used to detect the type of bond present and functional groups [15].
In vitro Anti-bacterial and Anti-Biofilm Efficacy Assessment
Antibacterial activity of extracted bio-surfactant: The following clinical pathogens were employed to test the antibacterial efficacy of extracted bio-surfactant, Enterococcus faecalis, Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa and Staphylococcus aureus. Sterile distilled water used to adjust the turbidity of test culture suspension according to McFarland standard of 0.5. Well diffusion method and Muller Hinton Agar medium were used for the assessment of antibacterial activity [16]. Hundred microliters of the culture suspension swabbed over the medium with sterile cotton swab and 6 mm sized wells were pierced using cork-borer. The plates were sealed and incubated at 37°C for 24 hr and the zone of inhibition was measured with zone scale [17].
Anti-biofilm efficacy analysis of extracted bio-surfactant: About 100 μL of biofilm forming, pathogenic E. coli was added to 96-well plate and incubated with lid at RT (37°C) for 24 hr in an orbital shaker. After the completion of biofilm forming assay, 100 μL of extracted bio-surfactant added into the same wells containing biofilm [18]. The plate was incubated at 37°C for 2 hr. In two different wells, 1% of Sodium Dodecyl Sulfate (SDS) loaded as positive control and E. coli culture used as negative control. Planktonic cells discarded from the suspension and again washed three times with sterile saline water [19]. The remaining cells attachment was fixed with methanol and dried. Cells were stained with 2% crystal violet stain for 15 min and washed. Then 150 μL of 33% glacial acetic acid was added to solubilize cells. In another 96-well plate 100 μL of solubilized suspension taken and optical density was measured at 600 nm using micro-plate reader. The percentage of inhibition was calculated (Formula 2) [20, 21].
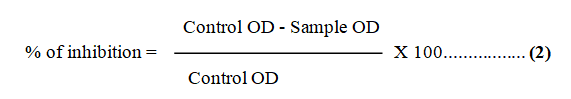
Isolation and Screening of Bio-Surfactant Producers
Oil containing sediment sample was collected from marine shores of Ennore and the same was subjected to serial dilution up to six folds. From the last two dilutions (10-5 and 10-6) eight different healthy individual strains were obtained and they were adapted to different screening methods for the isolation of novel BS producer. A red pigment producing bacterial strain shown positive results to all screening methods, this strain was selected for further studies. A genus and species of the selected strain was identified as Serratia marcescens MT053078 by 16S rRNA sequencing analysis and the new strain was submitted to NCBI (Figure 1).
Figure 1: Red pigment producing Serratia marcescens MT053078 isolated by serial dilution.
Genotypic and Phenotypic Characterization
The individual colony of the novel strain S. marcescens MT053078 is motile, rod shaped, pigment producer and the cells were observed by compound microscope to be grams' negative. An antibiotic susceptibility test was recorded by measuring the zone of clearance by zone scale and values are tabulated. From the table we have concluded that the strain S. marcescens MT053078 is highly sensitive to 10 µg of gentamicin and totally resistance to 10 µg of ampicillin (Table 2 and Figure 2). DNase test was observed to be positive by zone of clearance on DNase agar medium (Figure 3). Additionally, the isolated strain provided positive results to some more enzymatic tests such as gelatin hydrolysis, lipase and catalase.
| Antibiotics | Zone of clearance (in mm) |
|---|---|
| SD016-1CT Gentamicin (Gen10) | 11 |
| SD002-1CT Ampicillin (Amp10) | 0 |
| SD037-1CT Tetracycline (TE30) | 3 |
| SD021-1CT Nalidixic acid (NA30) | 7 |
| SD006-1CT Chloramphenical (C30) | 8 |
| SD003-1CT Streptomycin (S10) | 4 |
Table 2: Antibiotic susceptibility test of Serratia marcescens MT053078.
Figure 2: Antibiotic susceptibility test of Serratia marcescens MT053078.
Figure 3: Confirmation of DNase production by Serratia marcescens MT053078.
Optimization and Extraction
The rapid cell proliferation and maximum secondary metabolites production were strongly influenced by making minor changes in the physical and chemical parameters of the growth medium. The high yield was measured by emulsification index during medium optimization. More amount of BS was produced at 40°C (Figure 4) in neutral pH (7) (Figure 5) at 150 rpm (Figure 6) with 1 % NaCl (Figure 7). S. marcescens MT053078 is a facultative anaerobe which can survive without aeration, so they were produced maximum BS even in static condition (Figure 8) with 80 % of emulsification index.
Figure 4: Effect of temperature on production of bio-surfactant by Serratia marcescens MT053078.
Figure 5: Effect of pH on production of bio-surfactant by Serratia marcescens MT053078.
Figure 6: Effect of rpm on production of bio-surfactant by Serratia marcescens MT053078.
Figure 7: Effect of salt on production of bio-surfactant by Serratia marcescens MT053078.
Figure 8: Effect of static condition on production of bio-surfactant by Serratia marcescens MT053078.
The carbon source, fructose shown best EI with the (Figures 9 and 10) concentration of 0.2%, whereas nitrogen source (Figures 11 and 12) (casein enzymic hydrolysate) exhibited best EI with 1%. Edible oils used as enrichment, for this castor oil and mg from metal ions used were (Figures 13 and 14) shown better EI with maximum BS production. Apart from these, incubation period was checked for all the selected parameters together, the strain S. marcescens MT053078 can produce more BS at 48 hrs of incubation (Figure 15) than 24 hrs.
Figure 9: Effect of carbon sources on production of bio surfactant by Serratia marcescens.
Figure 10: Different concentrations of fructose on production of bio-surfactant by Serratia marcescens.
Figure 11: Effect of nitrogen sources on production of bio-surfactant by Serratia marcescens MT053078.
Figure 12: Different concentrations of casein enzymic hydrolysate on production of bio-surfactant by Serratia marcescens MT053078.
Figure 13: Effect of edible oils on maximum production of bio-surfactant by Serratia marcescens MT053078.
Figure 14: Effect of metal ions on maximum production of bio-surfactant by Serratia marcescens MT053078.
Figure 15: Effect of incubation period on maximum production of bio-surfactant by Serratia marcescens MT053078.
Crude BS was obtained from mass cultivation by acid precipitation and liquid-liquid extraction methods. The organic layer was separated out and kept in a desiccator with sodium sulphite for overnight to remove the water molecules by complete evaporation. Finally the end product was purified with sodium bicarbonate and freeze dried (Figure 16).
Figure 16: Organic layer formation during bio-surfactant purification: A) Crude bio-surfactant; B) Purified bio surfactant.
Characterization of Bio-Surfactant
The purified powder BS was initially characterized by TLC to confirm the purity and to identify the special group of BS. The separated spot by mobile phase was visualized in UV chamber and ninhydrin reagent gently sprayed over the TLC plate (Figure 17). Formation of pale purple colour spot confirmed that the extracted BS belongs to lipopeptide group. The FT-IR spectrum (Figure 18) for the purified BS shown peaks that corresponded to O-H, C=C or C=O and C-H functional groups.
Figure 17: TLC of purified bio-surfactant (a) Sample in TLC chamber (b) Compound shows fluorescence in UV chamber (c) Ninhydrin sprayed over TLC plate.
Figure 18: FTIR analysis of purified bio-surfactant of Serratia marcescens MT053078.
Applications of Bio-Surfactant
In vitro antibacterial activity: The zone of clearance was observed around the positive control gentamicin as well as around the BS sample followed by the desired incubation period (Figure 19). The clearance came up absent around the negative control. Zone scale was used to measure the inhibition zone in mm and the results are shown in Table 3. At the employed concentration, the extracted bio-surfactant had good activity. If the concentration is boosted, the activity can be dramatically enhanced.
Figure 19: Antibacterial activity of bio-surfactant extracted from Serratia marcescens MT053078 against Escherichia coli.
| Test organisms | Zone of Inhibition (mm) | ||
| Negative Control |
Sample | Gentamicin | |
| Enterococcus faecalis | 0 | 06 ± 0.02 | 05 ± 0.06 |
| Escherichia coli | 0 | 08 ± 0.04 | 06 ± 0.08 |
| Klebsiella pneumonia | 0 | 06 ± 0.04 | 09 ± 0.05 |
| Pseudomonas aeruginosa | 0 | 07 ± 0.06 | 07 ± 0.06 |
| Staphylococcus aureus | 0 | 05 ± 0.05 | 08 ± 0.08 |
Table 3: Antibacterial activity of extracted bio-surfactant against clinical pathogens by well diffusion method.
The maximum zone of clearance was 8 mm for E. coli and the lowest was 5 mm for S. aureus. In respect to E. faecalis, the minimum zone of clearance was noted as 6 mm. It is abundantly obvious from the results that the BS derived from S. marcescens MT053078 had a greater effect on clinical pathogens than the positive control employed.
In vitro anti-biofilm activity: At the end of anti-biofilm assay, the solubilized cell suspensions were absorbed by micro-plate reader (ELISA) at 600 nm. Table 4 shows the notable activity of bio-surfactant against E. coli and they were used in-order to compute the percentage of inhibition using the (Figure 20) standard formula, which was already stated in methodology section (10.2). The inhibition of growth was observed and results clearly demonstrate that the microbial metabolite displayed prominent activity against E. coli. Based on the results, BS had a notable and very high biofilm dispersal efficacy of about 81.30 % compared to the positive control employed (Figure 21).
| E. coli | A | B | C | Mean | X | % of dispersal | |
|---|---|---|---|---|---|---|---|
| Control | 0.107 | 0.108 | 0.107 | 0.107 | Control-Sample | X/Control x 100 | |
| SDS | 0.021 | 0.022 | 0.020 | 0.021 | 0.107-0.021=0.086 | 80.37% (SDS) | |
| Sample | 0.022 | 0.019 | 0.021 | 0.020 | 0.107-0.020=0.087 | 81.30% (BS) |
Table 4: In vitro anti-biofilm activity of extracted bio-surfactant against Escherichia coli biofilm.
Figure 20: Anti-biofilm activity of bio-surfactant extracted from Serratia marcescens MT053078.
Figure 21: Percentage of biofilm dispersion using bio-surfactant.
The maximum bio-surfactant production was succeeded by making mild changes in the culture conditions. The gram negative and motile S. marcescens MT053078 can rapidly multiply at 400°C of optimum temperature with neutral pH. The study revealed that the maximal production of BS was obtained at 48 hrs of incubation in aerobic and anaerobic condition. The red pigment and excess BS (two-in-one) producing potential of S. marcescens MT053078 utilized for significant applications. The antibacterial activity results made it abundantly clear that the bio-surfactant had good impact on clinical pathogens. An anti-biofilm activity result was compared with the positive control used; bio-surfactant had a remarkable and extremely high biofilm dispersal efficacy of 81.37%. An attempt to be taken on bioremediation of crude oil/textile effluent with enzymatic assay and extraction of organic dye.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Vijayaraj R (2025) Extraction of Antibacterial Surface-Active Molecule from Serratia marcescens MT053078: Characterization and Biological Evaluation. Eur Exp Bio. 15:39.
Copyright: © 2025 Vijayaraj R. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.