Research Article - (2018) Volume 4, Issue 1
Li Cai*, Scott Hood, Eddie Kallam, Danny Overman, Kaitlyn Barker, Dylan Rutledge, Jordan Riojas, Carissa Best, Marcia Eisenberg and Lauren Kam-Morgan
Center for Molecular Biology and Pathology, Laboratory Corporation of America Holdings, Research Triangle Park, North Carolina, USA
*Corresponding Author:
Li Cai
Center for Molecular Biology and Pathology
Laboratory Corporation of America Holdings
Research Triangle Park, North Carolina, USA
Tel: 9196108243
E-mail: cail@labcorp.com
Received Date: March 06, 2018; Accepted Date: March 14, 2018; Published Date: March 19, 2018
Citation: Cai L, Hood S, Kallam E, Overman D, Barker K, et al. (2018) Epi proColon®: Use of a Non-Invasive SEPT9 Gene Methylation Blood Test for Colorectal Cancer Screening: A National Laboratory Experience. J Clin Epigenet 4:7. doi: 10.21767/2472-1158.100092
The Epi proColon® test is the first and only FDA-approved molecular blood test for colorectal cancer (CRC) screening. The high-sensitivity Real-Time PCR method detects a hypermethylated promoter region of the Septin 9 gene (SEPT9) shown to be associated with colorectal cancer when present in cell-free plasma. Following established protocols for new test assessments, the Epi proColon® test validation results were found to be comparable to the manufacturer’s stated performance characteristics. With familiar PCR methodology and minimal operator to operator variability, the test has been easily integrated into clinical molecular laboratory workflows. To accommodate our standardized process logistics, we validated the substitution of three 4 mL EDTA Vacutainer® collection tubes (Becton Dickinson) for the manufacturer-recommended single 10 mL tube for use in all patient specimen collection centers. By mid-year 2017, 2,238 Epi proColon® tests were ordered by our clinical provider-base for individuals of recommended screening ages 50-74. Positivity rates in this cohort were comparable to published clinical trial rates for the test. For those people who remain resistant to other methods of CRC screening, the availability of a blood test that may be conveniently drawn at a local community collection center may help surmount barriers of nonparticipation. This report summarizes LabCorp’s validation, implementation and clinical experience for on-boarding and offering the Epi-proColon test to healthcare providers and patients.
Keywords
Septin genes; BisDNA; Promoter region; Epi-proColon
Introduction
As the third leading cause of global cancer mortality, colorectal cancer (CRC) attributed nearly 700,000 deaths worldwide in 2012 and 50,000 in the United States in 2017 [1,2]. Despite the significant promotional efforts made by cancer and advocacy societies and organizations who publish recommendations and guidelines that advocate strongly for colorectal cancer screening— U.S. screening rates have continued to hover around 63% [3]. As a result, a third of the eligible population remains unscreened, leaving little opportunity to find cancer early in those who avoid participating in CRC screening programs. The difference between an early versus a late stage diagnosis is significant in terms of treatment success, survival and cost for cancer care. Currently, about 60% of CRC cases are diagnosed in later stages of cancer, therefore, a significant cost burden is associated with the diagnosis and treatment of later stage cancer [4,5]. Further, in a 2016 study that analyzed CRC diagnoses in a large unscreened population, the analysis found that 43% of new cases were directly attributed to those not participating in any type of a screening program [6].
A broad base of evidence supports the benefits of screening just as it reports the many reasons from non-compliance. Individualized personal patient barriers as well as the traditional endoscopic and stool-based methodologies are among the obstacles reported to impede participation. Blood sampling, as one of the most common and patient-accepted methods of testing, has demonstrated the potential to overcome personal barriers and encourage screening participation [7,8]. The Epi proColon® test is the first and only blood test to gain FDA approval (April, 2016) for colorectal cancer screening.
Background
SEPT9 Gene hypermethylation
As a normal process, DNA methylation is one of the most commonly occurring epigenetic events and vital components of many genetic processes including gene expression and transcription [9]. Alterations to normal DNA methylation patterns during early carcinogenesis make these events valuable markers for the early detection of cancer [10,11]. As a highly recognized alteration associated with a variety of human cancers, DNA hypermethylation typically occurs at CpG islands located in the promotor region of a gene, inactivating tumor suppressor genes and silencing transcription [10,12,13].
The SEPT9 gene encodes the Septin 9 protein, a member of a highly conserved family of GTP-binding proteins that function in key regulatory and cellular processes [14,15]. Located at chromosome 17q25.3 in human cells, SEPT9 is the most complex of the 13 septin genes containing 18 distinct transcripts encoding 15 isoforms [12,16]. SEPT9 plays important roles in actin dynamics, angiogenesis, bacterial autophagy, cell motility, cell proliferation, cell shape, cytokinesis, microtubule regulation, vesical targeting and exocytosis [12,15,16]. Abnormal or no expression of the SEPT9 gene critically affects cytokinesis, a key feature in CRC carcinogenesis [12,17].
Clinical significance of SEPT9 gene hypermethylation in CRC
CRC is both a genetic and epigenetic disease [11,18]. A complex set of epigenetic mechanisms regulate gene expression in both normal and cancerous tissue [13]. Aberrant DNA methylation, the most studied of these mechanisms in CRC, contributes to its heterogeneity and can be identified by unique methylated gene signatures [18]. Hypermethylation occurring in the promotor region of the SEPT9 _v2 transcript at CpG island 3 (CGI3) has been highly correlated with CRC carcinogenesis as well as discriminating cancer from non-cancerous colonic mucosa, Figure 1 [12,19]. Epigenetic silencing of the SEPT9 gene by promoter methylation in cell-free plasma has been shown to be a biomarker for CRC [12,16,17]. Using high-sensitivity Real-Time PCR methods, hypermethylated SEPT9 DNA signatures can be detected in cell-free plasma [12,19-27]. As an early event and hallmark of human cancers, aberrant DNA methylation biomarkers have clinical roles in screening, diagnosis, prognosis and therapeutic response [11,28].
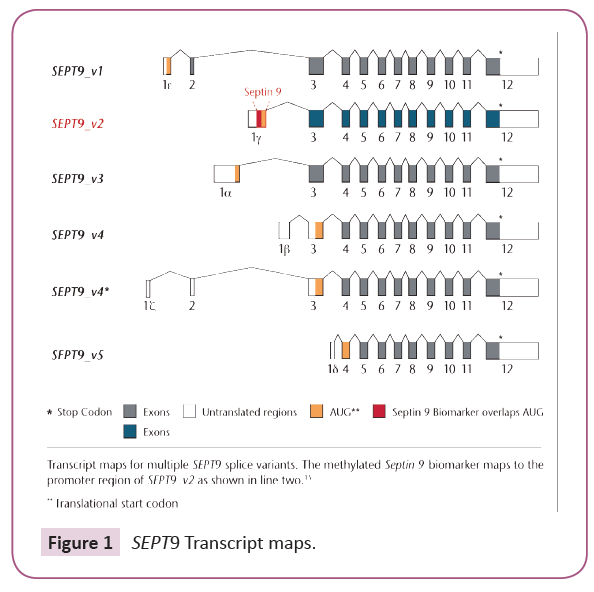
Figure 1: SEPT9 Transcript maps.
The Epi proColon Test®
The Epi proColon test is a qualitative in vitro diagnostic molecular method for the detection of methylated SEPT9 DNA (Septin 9) in EDTA plasma derived from patient whole blood specimens. Methylation of this target DNA sequence in the promoter region of the SEPT9_v2 transcript has been associated with the occurrence of CRC [9], Figure 1. This test is indicated to screen adults of either sex, 50 years or older, defined as average risk for CRC, who have been offered and have history of not completing CRC screening. People with positive Epi proColon® test results are referred for diagnostic colonoscopy to confirm the presence of CRC.
Methods
Setting
This study was designed and conducted by the LabCorp Specialty Testing Group at the Center for Molecular Biology and Pathology (Laboratory Corporation of America, Research Triangle Park, NC) to evaluate and validate the Epi proColon® blood test for the purpose of adding the molecular method as an offering in the test menu (Site One). Epigenomics AG (Berlin, Germany) served as Site Two, a comparator site for accuracy studies.
Plasma specimens
Accuracy and precision studies: Accuracy and precision were determined using K2EDTA plasma samples of known positive methylated SEPT9 status and healthy donor samples of presumed negative methylated SEPT9 status. Samples were derived from participant K2EDTA whole blood specimens and plasma prepared within four hours from the time of collection as required by the Epi proColon® Instructions for Use (IFU) [29]. Samples were prepared and stored at -70°C prior to testing.\
4 mL EDTA collection tube validation studies: These studies used plasma derived from three 4 mL and 10 mL whole blood K2EDTA collection tubes from healthy donors of presumed negative methylated SEPT9 status. Whole blood specimens were processed to plasma within four hours from the time of collection as required by the Epi proColon® IFU and stored at -70°C prior to testing.
Test Methodology
Following the Epi proColon® IFU, cell-free DNA was isolated from K2EDTA plasma using two centrifugation steps, Figure 2. Using the Epi proColon® Plasma Quick Kit, 3.5 mL of cell-free plasma DNA was bound on DNA-binding magnetic beads. Beads were then captured, washed and purified DNA was eluted. Next, the purified DNA was treated with a high concentration of ammonium bisulfite under denaturing conditions to generate bisulfite-converted DNA (BisDNA). The BisDNA was re-purified using magnetic beads, then assayed using the Epi proColon® Sensitive PCR Kit. Duplex Real-time PCR was performed on the Applied Biosystems® 7500 Fast Dx to detect the target methylated form of Septin 9 DNA.
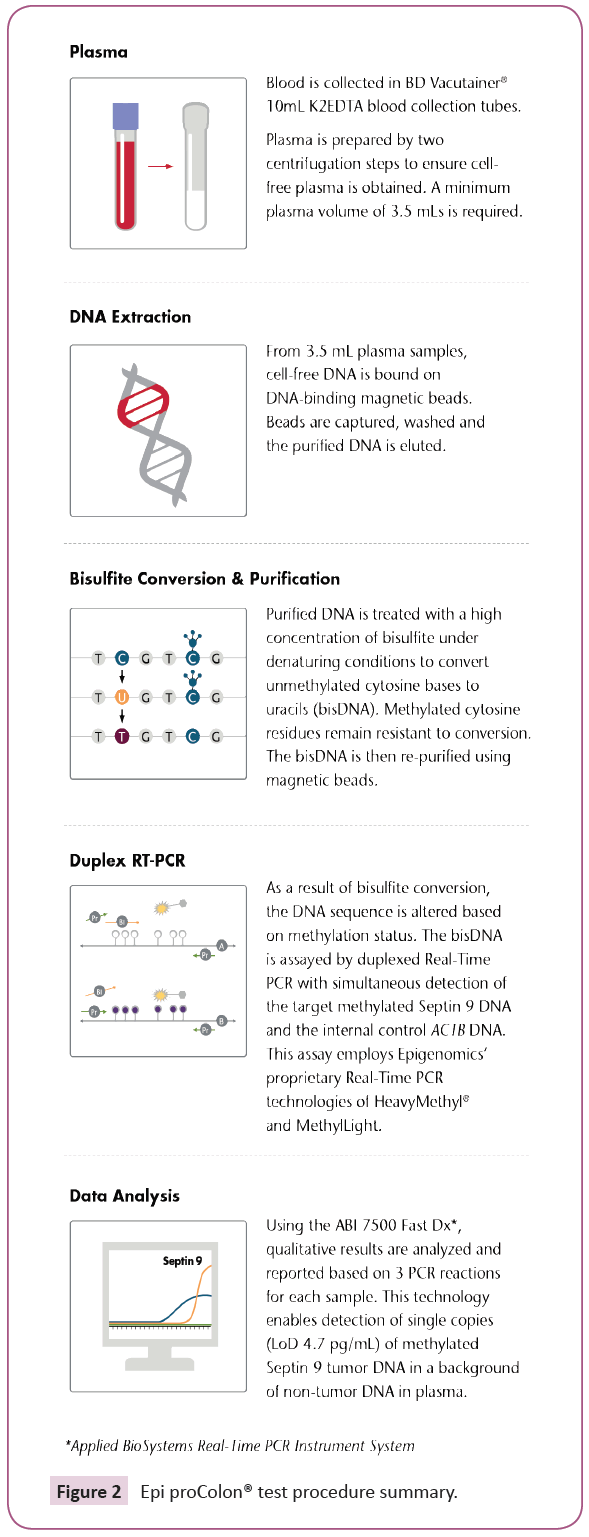
Figure 2: Epi proColon® test procedure summary.
A duplexed ACTB (β-actin) DNA was co-amplified to monitor for adequate DNA concentration, sample preparation and sample quality. Using the Epi proColon® Control Kit, external controls were processed with every run to ensure procedural competency and validity of test results. The qualitative test results were based on the three PCR reactions (Table 1). Positivity was determined using an algorithm whereby at least one of three PCR replicates tests had both a valid ACTB and positive Septin 9 result. Negativity was determined when all three PCR replicates had a valid ACTB and all three Septin 9 PCR replicates were negative. The Epi proColon® test was invalid in all other cases.
| Control result | Determination | Septin 9 result | ACTB result | Invalid result |
|---|---|---|---|---|
| Valid positive control result | PCR 1, 2, 3 | Ct ≤ 41.1 | Ct ≤ 29.8 | No positive and at least 1 of 3 invalid PCR replicate |
| Valid negative control result | PCR 1, 2, 3 | No Ct result (undetermined) |
Ct ≤ 37.2 | At least 1 of 3 invalid PCR replicate |
Table 1: Validity of Epi proColon® control results and interpretation.
4 mL Blood collection tube validation
LabCorp clinical laboratories and remote collection sites use three 4 mL K2EDTA Vacutainer collection tubes (Becton Dickinson). A validation study was conducted to ensure adequate plasma volume was obtained and test performance was not affected by the tube substitution. A minimum plasma volume of 3.5 mL was required to perform the Epi proColon® test, therefore, three 4 mL K2EDTA tubes were required to obtain an adequate volume of plasma for testing.
Two sets of blood specimens, each set containing three 4 mL and one 10 mL K2EDTA blood collection tubes, were collected from 36 healthy donors with presumed negative methylated SEPT9 status. Whole blood specimens were processed to plasma within four hours from the time of collection. Plasmas from each group of three 4 mL K2EDTA collection tubes were pooled into a single transport cryotube and frozen to simulate the collection center protocol. Plasma pools were prepared in the same manner for the 10 mL K2EDTA collection tubes. Frozen plasma aliquots were shipped to testing Site Two for analysis. All testing runs were performed by a single operator on the ABI 7500 Fast Dx and included positive and negative external controls and an ACTB internal control.
Accuracy and precision
The validation plan included studies designed to evaluate accuracy and inter-assay and intra-assay precision. Accuracy and precision were determined using K2EDTA plasma samples of known positive methylated SEPT9 status and healthy donor samples of presumed negative methylated SEPT9 status. For accuracy studies, a total of twenty plasma samples were evaluated and results compared for positive and negative agreement between the two testing sites, using PCR triplicates as required for the Epi proColon® test method.
For precision studies, plasma pools were used to accommodate the sample volume required to analyze samples in multiple runs of three PCR replicates per sample. Three pools of plasma of known positive methylated SEPT9 status and three pools of plasma of presumed negative methylated SEPT9 status were used. Each pool was comprised of five status-associated plasmas.
Clinical experience patient testing
Following the Epi proColon test instructions for specimen collection, transport and processing, patient whole blood specimens (K2EDTA) were collected and processed to plasma within the required four-hour timeframe, Figure 3. We analyzed test positivity in a series of 2,238 screening age-eligible patients whose testing was performed as a standard clinical referral. Patients were de-identified, and analyzed in aggregate as well as by patient age.
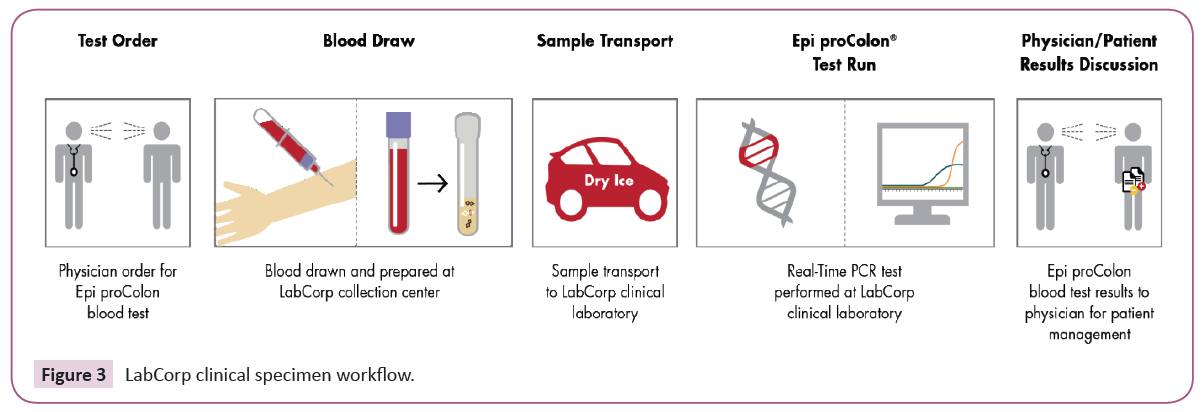
Figure 3: LabCorp clinical specimen workflow.
Results
4 mL Collection tube validation
Combining plasma aliquots from the three 4 mL blood tubes achieved greater than the required 3.5 mL of plasma in all cases. Results generated from the 4 mL and 10 mL K2EDTA Vacutainer collection tube sizes demonstrated high concordance for both the known positive and presumed negative samples. Of the 72 presumed healthy samples tested and compared, three samples obtained positive results in each group of 36 for a combined positivity rate of 8.3% (6/72) and negative agreement of 91.7% (66/72). No change or increase in Septin 9 positivity rate was observed using either type of collection tube in the tested population. Cycle threshold (Ct) values for the ACTB internal control assay for the 4 mL tubes were slightly lower than for the 10 mL tubes, indicating more DNA in the plasma with a median difference of 0.15 Ct. Since the study was a simple, repeated measurement of ACTB variability, the standard deviation was 0.45 Ct and comparable to the variation observed in the Epi proColon® assay’s reproducibility studies.
Accuracy
Known positive methylated SEPT9 status samples: The analysis of the ten plasma specimens with known positive methylated SEPT9 status showed 100.0% (20/20) concordance of positivity and agreement between the two sites (Table 2).
| Testing sites | Known SEPT9 positive status | Presumed SEPT9 negative status | Septin 9 positive/negative results | % Positive agreement | % Negative agreement |
|---|---|---|---|---|---|
| Site 1, n=20 | 10 | 10 | 10/7 | 100.0 | 80.0 |
| Site 2, n=20 | 10 | 10 | 10/8 | 100.0 | 70.0 |
| Combined, n=40 | 20 | 20 | 20/15 | 100.0 | 75.5 |
Combined accuracy for both sites=87.5% (35/40)*
*Samples of Known positive methylated SEPT9 status and healthy donors with presumed negative methylated SEPT9 status
Table 2: Accuracy.
Presumed negative methylated SEPT9 status samples: The analysis of the ten specimens from healthy donors of presumed negative methylated SEPT9 status were 75.0% (15/20) concordant for both sites combined, and 70.0% (7/10) and 80.0% (8/10) individually at Site One and Two laboratories, respectively. Negative samples resulting in a positive interpretation were based on one of three PCR replicates with a positive Septin 9 Ct value.
Intra-assay precision
Three plasma pools of known positive methylated SEPT9 samples and three plasma pools of presumed negative methylated SEPT9 samples were tested in triplicate, within one run, for intra-assay precision (Table 3). There was 100% (9/9) concordance for all methylated SEPT9 positive specimens and 77.8% (7/9) for methylated SEPT9 negative specimens. For two discordant negative samples, only one of three PCR replicates had a positive methylated SEPT9 Ct result. Overall concordance among the eighteen tested specimens was 88.9% (16/18).
| Inter and intra run 1 | Pool ID | ACTB Ct | Septin 9 Ct | Result | ||||
|---|---|---|---|---|---|---|---|---|
| PCR1 | PCR2 | PCR3 | PCR1 | PCR2 | PCR3 | |||
| Negative pool 1 | A | 27.7 | 28.4 | 28.2 | UND | UND | UND | Negative |
| B | 28.2 | 28.5 | 28.6 | UND | UND | UND | Negative | |
| C | 27.3 | 28.5 | 28.5 | UND | UND | UND | Negative | |
| Negative pool 2 | A | 28.4 | 28.8 | 28.3 | UND | UND | UND | Negative |
| C | 28.4 | 28.6 | 28.6 | UND | UND | UND | Negative | |
| E | 28.0 | 28.0 | 27.7 | UND | UND | 39.7 | Positive | |
| Negative pool 3 | A | 28.1 | 27.8 | 28.6 | UND | 37.6 | UND | Positive |
| B | 28.5 | 27.7 | 28.5 | UND | UND | UND | Negative | |
| C | 28.6 | 28.5 | 28.5 | UND | UND | UND | Negative | |
| Positive control | 27.1 | 27.2 | 27.5 | 35.5 | 34.3 | 34.6 | Positive | |
| Negative control | 30.6 | 30.5 | 30.6 | UND | UND | UND | Negative | |
*Samples from healthy donors with presumed negative methylated SEPT9 status
Table 3: Intra-assay and inter-assay precision: Run 1.
Inter-assay precision
Three plasma pools of known positive methylated SEPT9 samples and three plasma pools of presumed negative methylated SEPT9 samples were tested in triplicate, in three runs, for inter-assay precision (Tables 3-5). The intra-assay precision run was used as one of the 3 independent inter-assay runs.
| Inter-run 2 | Replicate | ACTB Ct | Septin 9 Ct | Result | ||||
|---|---|---|---|---|---|---|---|---|
| PCR1 | PCR2 | PCR3 | PCR1 | PCR2 | PCR3 | |||
| Positive 1 | D | 23.4 | 23.4 | 23.4 | 33.9 | 33.9 | 34.3 | Positive |
| Positive 2 | D | 24.2 | 24.2 | 24.3 | 35.3 | 35.9 | 35.6 | Positive |
| Positive 3 | D | 23.8 | 23.8 | 23.9 | 36.2 | 35.7 | 35.4 | Positive |
| Negative 1 | D | 28.3 | 28.3 | 28.2 | UND | UND | UND | Negative |
| Negative 2 | B | 28.4 | 28.3 | 28.5 | UND | UND | UND | Negative |
| Negative 3 | D | 28.6 | 28.3 | 28.3 | UND | UND | UND | Negative |
| Positive control | 28.2 | 28.4 | 28.6 | 36.1 | 35.7 | 38.8 | Positive | |
| Negative control | 32.0 | 32.0 | 31.6 | UND | UND | UND | Negative | |
*Samples of known positive methylated SEPT9 status and healthy donors with presumed negative methylated SEPT9 status
Table 4: Inter-assay precision: Run 2.
| Inter-run 3 | Replicate | ACTB Ct | Septin 9 Ct | Result | ||||
|---|---|---|---|---|---|---|---|---|
| PCR1 | PCR2 | PCR3 | PCR1 | PCR2 | PCR3 | |||
| Positive 1 | D | 23.4 | 23.4 | 23.4 | 33.9 | 33.9 | 34.3 | Positive |
| Positive 2 | D | 24.2 | 24.2 | 24.3 | 35.3 | 35.9 | 35.6 | Positive |
| Positive 3 | D | 23.8 | 23.8 | 23.9 | 36.2 | 35.7 | 35.4 | Positive |
| Negative 1 | D | 28.3 | 28.3 | 28.2 | UND | UND | UND | Negative |
| Negative 2 | B | 28.4 | 28.3 | 28.5 | UND | UND | UND | Negative |
| Negative 3 | D | 28.6 | 28.3 | 28.3 | UND | UND | UND | Negative |
| Positive control | 28.2 | 28.4 | 28.6 | 36.1 | 35.7 | 38.8 | Positive | |
| Negative control | 32.0 | 32.0 | 31.6 | UND | UND | UND | Negative | |
**Samples of known positive methylated SEPT9 status and healthy donors with presumed negative methylated SEPT9 status
Table 5: Inter-assay precision: Run 3.
For the three independent inter-assay runs, there was 100% (9/9) concordance for all methylated SEPT9 positive specimens and 77.8% (7/9) for methylated SEPT9 negative specimens. For the two discordant samples, one of three PCR replicates had a positive methylated Septin 9 Ct result, rendering the interpretation of a positive result. Overall concordance among the eighteen tested specimens was 88.9% (16/18).
Combined intra and inter-assay precision data
In total, six plasma pools, three positive and three negative, comprised of five samples per pool, were assayed five times for a total of thirty results. Combined positive percent agreement for inter and intra-assay methylated SEPT9 positive sample pools showed 100% concordance (15/15) compared to 98.0% (129/132) for sample pools of known positivity, as stated in the Epi proColon® IFU. Combined negative percent agreement in presumed negative methylated SEPT9 plasma samples showed 86.7% concordance (13/15), an improvement over the 75.0% (27/36) negative percent agreement observed in healthy donors as stated in the Epi proColon® IFU. Overall percent agreement for all six plasma pools of positive and negative samples evaluated for precision resulted in a 93.3% (28/30) concordance, which was comparable to the manufacturer’s stated 93.0% (156/168) (Table 6).
| Known SEPT9 positive status | Presumed SEPT9 negative status | Septin 9 positive/negative results | % Positive agreement | % Negative agreement | Overall agreement | |
|---|---|---|---|---|---|---|
| Intra-precision, n=18 | 9 | 9 | 18/16 | 100.0 | 77.8 | 88.9 |
| Inter-precision, n=18 | 9 | 9 | 18/16 | 100.0 | 77.8 | 88.9 |
Combined intra and inter assay precision overall concordance=93.3% (28/30)*
*Samples of known positive methylated SEPT9 status and healthy donors with presumed negative methylated SEPT9 status
Table 6: Intra-assay and inter-assay precision, combined.
Clinical experience data
Based on the validation of the test’s performance characteristics, the Epi proColon® was offered as a clinical test at LabCorp. Of the 2,238 plasma specimens tested, negative, positive and invalid results were 73.9%, 23.6% and 2.5%, respectively, Figure 4. Among the samples with positive results, 67.8% were determined as positive based on one of three positive replicates, 19.1% on two of three positive replicates and 13.1% on three of three positive replicates. Results could not be obtained on an average of 2.5% of samples due to low amounts of recoverable DNA. The positive detection rate was similar to the manufacturer’s rate and there were no observed differences based on gender.
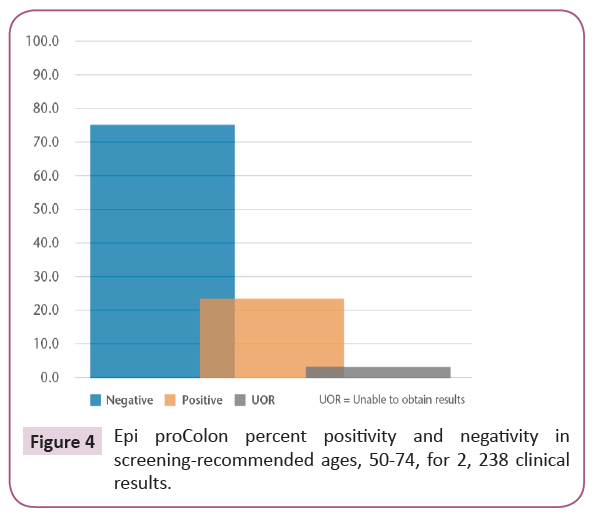
Figure 4: Epi proColon percent positivity and negativity in screening-recommended ages, 50-74, for 2, 238 clinical results.
Discussion
The Epi proColon® test, as the first FDA-approved blood test for colorectal cancer screening, offers a non-invasive method more likely to be accepted in the population who continues to resist screening. To date, access to healthcare and conveniences associated with getting tested have been major barriers that have prevented screening. From a patient perspective, personal factors that cause patients to be unwilling or unable to accept the more traditional endoscopic and stool methods may be overcome by this common method of blood testing. From an access point of view, offering this blood test through a large national clinical laboratory with widely-dispersed community collection centers provides a testing convenience that serves to enhance screening opportunities.
Overall, the Epi proColon® test validation demonstrated results for accuracy and precision comparable to the assay’s established performance characteristics reported in the IFU. The test showed minimal intra or inter operator-to-operator, run-to-run or lot-lot variability. Accuracy studies using samples of known positive methylated SEPT9 and presumed negative methylated SEPT9 status showed 100%, 75%, and 87.5% concordance for positive, negative and overall agreement of results at the two testing sites. Precision studies were performed at a single site using three pools of known positive SEPT9 methylation status and three plasma pools of presumed negative methylation SEPT9 status from healthy donors. Both intra-assay and inter-assay precision studies showed 100%, 77.8% and 88.9% concordance for positive, negative and overall agreement of results.
Standard LabCorp protocols established for collection centers use 4 mL Vacutainer K2EDTA collection tubes throughout the country. To maintain current processing and logistics workflow, validation of 4 mL Vacutainer K2EDTA tubes was performed and demonstrated high positive and negative concordance versus the recommended 10 mL tube. Both the 4 mL and 10 mL collection tubes showed comparable 8.3% positivity and 91.7% negative agreement rates in presumed negative methylated SEPT9 samples.
Based on total results of the evaluation, the Epi proColon® SEPT9 DNA blood test was offered to the national clinical-referral base. A blood collection and handling protocol, developed for the collection centers, required collection of six 4 mL Vacutainer K2EDTA tubes to generate sufficient plasma for repeat testing, sample rejection criteria and sample storage and transport requirements. Additionally, the protocol included the processing of whole blood specimens to plasma within the required four-hour timeframe of collection and freezing plasma aliquots at -15°C to -25°C for transport to the LabCorp laboratory for testing.
The LabCorp clinical laboratory processed 2,238 Epi proColon® tests submitted by clinical providers from patients of screening-age by mid-2017. Of these specimens from patients ages 50-74, 75.1% and 22.4% obtained negative and positive Epi proColon® test results, respectively. Due to limitations surrounding confidentiality, outcomes for patients obtaining positive results were unknown. Results could not be obtained in 2.5% of specimens due to insufficient recovery of plasma DNA. We found this rate comparable to exclusions found for other molecular methods such as FIT-stool DNA (2.2-3.8%) [30,31].
In the U.S., cancer organizations and professional societies recommend screening for individuals, ages 50-75, who are at average risk for colorectal cancer. For the 2,238 of the clinical specimens received for screening from individuals of screeningeligible- age, the overall positivity rate was 22.4%, comparable to the Epi proColon® test’s reported positivity rate [20].
Conclusion
The Epi proColon® test is a simple and robust, Real-Time PCR assay for molecular CRC screening of asymptomatic patients using cell-free plasma specimens. Following established protocols for new test assessments, the Epi proColon® validation results were comparable to the manufacturer’s stated performance characteristics. Familiar PCR methodology with minimal operator to operator variability enables easy test integration into clinical molecular laboratory workflows. To accommodate standardized nationwide process logistics for test use in collection centers, standard 4 mL EDTA Vacutainer® collection tubes were validated. Since on-boarding the Epi proColon® test, there has been a steady increase in utilization by the domestic clinical-referral base. As a large clinical laboratory with extensive national outreach capability, this test extends the ability to provide greater access to non-invasive CRC screening with a blood test option to providers and their patients. By mid-2017, 2,238 Epi proColon® tests were received and processed from the clinical referral base for individuals of recommended screening ages 50-74. For those people who remain resistant to other methods for CRC screening, the availability of a blood test that may be conveniently drawn at a local community collection center may help surmount barriers of non-participation.