Research Article - (2023) Volume 4, Issue 3
Received: 18-May-2023, Manuscript No. rgp-23-17314; Editor assigned: 23-May-2023, Pre QC No. rgp-23-17314 (PQ); Reviewed: 06-Jun-2023, QC No. rgp-23-17314; Revised: 11-Jul-2023, Manuscript No. rgp-23-17314 (R); Published: 17-Aug-2023, DOI: 10.21767/RGP.4.3.22
Background: Postaxial polydactyly is a congenital disorder of the limb abnormalities by posterior extra digits. The ZNF141 gene mutation in the N-terminal region were recently linked with PAP typeA. The ZNF141 gene have two domain C2-H2. Typically, C2-H2 domain comprise two cysteine in one chain and two Histidine in other chain. The missence mutation occur in chromosome 4P16.3 varient ID Q15928.1, result amino acid changes at 474 of threonine into isoleucine.
Methods: In this study a missense mutation determines by in Pakistani families which cause PAP type A. Whole exome sequencing identified by the missense mutation was carried out by the causative gene. Bioinformatics tools were used for confirmation of pathogenicity of the identified mutations.
Results: The novel missense mutations, c.1420C>T; p.T474I in ZNF141 gene were identified in Pakistani families, respectively. Whole exome sequencing confirmed co-segregation of the mutations with disease phenotypes in Pakistani families. In silico analysis of the sequence variants confirmed their pathogenicity.
Conclusion: The report of ZNF141 gene mutations in Pakistani families, which support the frequent involvement of ZNF141 sequence variants in obesity. The study provide molecular docking to the active binding site of ZNF141 protein with PAP type A.
PAP type A; Missense variants; Modelling; Docking; ZNF141
Polydactyly is the most commonly known as hyperdactyly or hexadactyly. polydactyly is the most congenital and foot deformity with an additional fingers or toe or any complex digital part [1]. In mature human being, there are 213 bones, excluding the sesamoids. Bones is a connective tissue that metabolically active to provide structural stability, movement, controlling and support [2]. Bones can be classified in different ways including by location shape and size. The long bones of the limb is upper arm and smaller bones of feet and hands are all tubular bones. Diaphysis is thick cortical bones, twin metaphysis is flared end and epiphyses contain anterior part of the bones. Polydactyly is the most severity in different affected individual. Some finger have no bones and some have bones but no joint and some of the completed functional finger. Polydactyly is classified in to three major groups [3]. Preaxial polydactyly, postaxial polydactyly and complex polydactyly. Postaxial polydactyly is further classified in to type A and type B. It is typically inherited in autosomal dominant trait in a diverse population. This disease is the most commonly in African peoples. It is most commonly caused by anterior-posterior in limb development [4]. It can affect 1.6-10.7 and 0.3-3.6 people per 1000 in live birth. The ZNF141 gene cause postaxial polydactyly type A disease. A sequence variant (c.1420 C>T; p.Thr474Ile) in the ZNF141 cause PAP typeA located on the chromosome 4p16.3. This mutation the threonine are converted into isoleucine. It is autosomal recessive which may affect the function of this protein in limb development. The ZNF141 gene have two domain C2-H2. Typically C2-H2 domain comprise two cystein in one chain and two Histidine in other chain. The ZNF141 gene contain 1422bp which encode 474 residues amino acid. The ZNF141 gene is expressed in spleen, pancrease, kidney, brain, liver and lungs. The ZNF141 gene sequence variant (c. 1420 C>T; p.Thr474Ile) to analyze it computationally by different bioinformatics tools.
Sequence Variant
To study the sequence variant which cause phenotypically postaxial polydactyly type A. A syndrome in human being the sequence variant was packed from the literature study of the ZNF141 gene. The target variant of ZNF141 gene protein FASTA sequence was retrieved from the NCBI database with accession ID:15928. The ZNF141 gene mutation was confirmed by the ensemble under the variant table [5]. Furthermore the affect of sequence variant on protein structure and function was confirmed the target variant through computational tools. SNAP2, Fathmm, Polyphen2, SIFT, predict SNP, I-Mutant to find the structural and functional consequence of variant (c.1420 C>T lead in to single amino acid p.Thr474Ile).
Phylogenetic Analysis
Phylogenetic as evolutionary tree to construct among ZNF141 gene were compared with 18 other closed related species as identified by NCBI-BLAST program to check the maximum resemblance and confirmation of related gene family in other species. The MEGA X programme used neighbour-joining techniques to establish the phylogenetic relationship between protein sequences, ensuring the statistical significance of the tree.
Structural Analysis and Validation
The ZNF141 gene 3D structure is not present in the PDB (Protein Data Bank). The FATSA sequence of ZNF141 gene was retrieved from NCBI databse. The structure was predicted through online server AlphaFold. The refining approach which has been tested CASP 10 method. Through Ramachandran plot and Errat Madhavan was generated 3D structure was validated to check the phi and psi angles. For more studying the validated 3D structure refine through rampage and galaxy refine tools.
Structure Mutagenesis and Comparison with Wild Type ZNF141
The 3-dimensional structure of the mutant protein from the wild type protein through chimera. The mutant structure was generated from the wild type protein. The chimera interface accepts user commands that are script-based, runs the commands, an has a graphical user interface. In the mutagenesis to demonstrate different rotamers in chimera. Each rotamers the mutation percentage valve which show how likely are the chances of each rotamers [6]. After that, the structure's potential clashes were cleared and eliminated.
Energy Minimization and Deleterious
The 3-dimensional structure energy minimization was carried out through swiss PDB. Muatant and wild type structure compared for energy minimization. To investigate the variant analysis using hope server, that will explore at the mutation change lying the novel structure. Hope server was used to draw residue to residue comparison.
Molecular Docking
For more studying, the ZNF141 gene were docking through different computational tools. The ligand was picked from ChemBle and design through autodock 1.5.7. The structural comparison of docked molecules was designed by Biovia discovery studio. To study how two or more molecular structure are fit together. The method through which a protein interacts with small molecule (ligand).
Sequence Variant Annotation
The ZNF141 gene sequence variant were retrieved from ensemble genome browser and uniprot databases. The ZNF141 gene identified a missense mutation (c.1420 C>T; p.Thr474Ile) located on the chromosome 4p16.3. The mutation of the results that the Threonine are converted in to Isoleucine. For more studying various online tools were tools for the structural and functional annotation of sequence variant. SNAP2 is affected the protein sequence with a score as =18% and 59% accuracy that the mutation can affect the disease. Predict SNP affect the disease with a score as 61%. Polyphen2 indicate the probably damaging with a score as 1.000. SIFT is affect the protein function with a score as 0.00 mutation can the disease. PROVEAN was deleterious that affect the protein structure with a score as -4.810. I-Mutant affect the protein structure stability with a score as -1.40.
Phylogenetic Analysis
Based on BLAST results, the phylogenetic tree was also constructed to confirm the conservation leptin with these 18 different species listed in Figure 1 belonging to the ZNF141 gene family as highly conserved [7]. The branch lenght show among the relation to which species are closely related to the human ZNF141 gene.
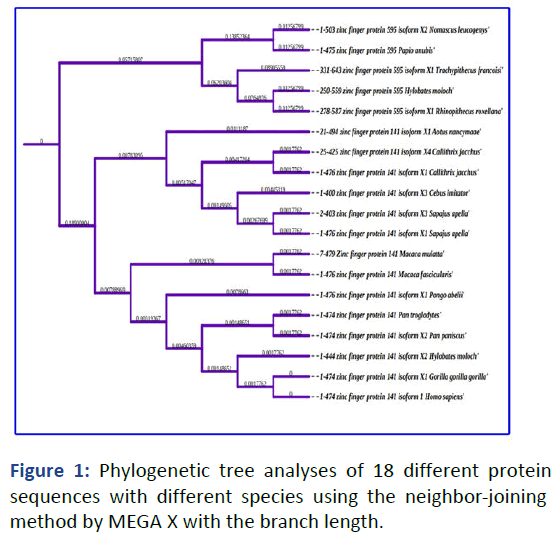
Figure 1: Phylogenetic tree analyses of 18 different protein sequences with different species using the neighbor-joining method by MEGA X with the branch length.
Secondary Structure Prediction and Mutagenesis
The secondary structure prediction was retrieved from PDBsum tool. There are 11 sheets, 11 beta hairpin, 22 strands, 8 beta bulges, 20 helices, 6 helix-helix interaction, 33 beta turn and 1 gamma (Figure 2). To further the secondary prediction to check the solvent accessibility through raptorx tool of wild type and mutant type. The result was consistent which depicted that decrease and increase solvent accessibility of ZNF141 gene causing postaxial polydactyly type A.
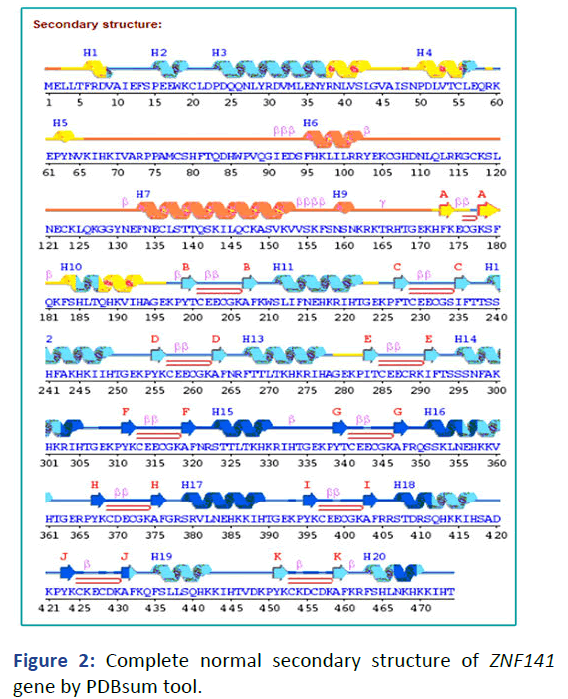
Figure 2: Complete normal secondary structure of ZNF141 gene by PDBsum tool.
Structural Analysis and Validation
The ZNF141 gene protein sequence was retrieved from ensemble genome browser. The 3-dimensional structure was obtained from online web based tool AlphaFold. The validation of 3-dimensional structure through different tools that are use for to check the protein properties for structure validation. Ramachandran plot and ERRAT were use for structure validation. Ramachandran plot show that 95.1% of residues are most favored region and 4.6% residues in allowed region and no residues fall in the outlier region. ERRAT show that the quality factor score as 90%. To further enhance the reliability that the 3-D structure refine through online tool galaxy refine.
Structure Mutagenesis and Comparison with Wild Type ZNF141
The mutant structure of ZNF141 was aligned with the wildtype ZNF141 structure, both structures were subjected to energy minimization prior to comparison [8]. The structural comparison revealed distinct differences in the residues between the two forms. Specifically, the side chains of the residues showed a notable change from threonine (wild-type) to isoleucine (mutant-type). Comparison of the residues demonstrated that the wild-type residues were smaller in size compared to the corresponding residues in the mutanttype structure (Figure 3).
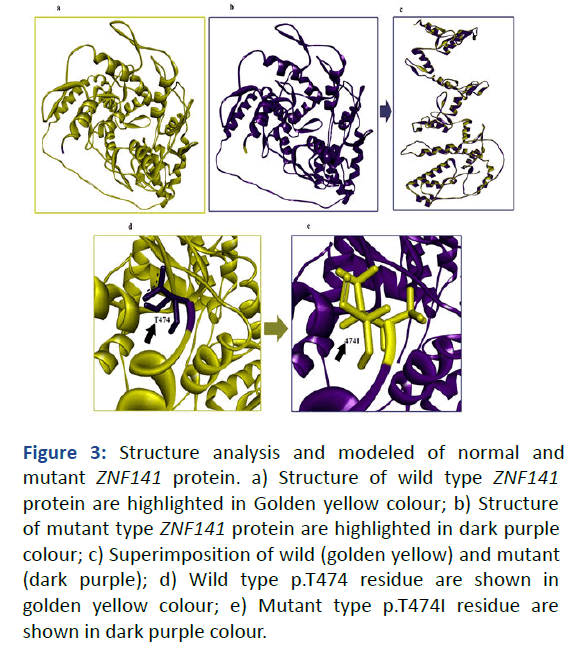
Figure 3: Structure analysis and modeled of normal and mutant ZNF141 protein. a) Structure of wild type ZNF141 protein are highlighted in Golden yellow colour; b) Structure of mutant type ZNF141 protein are highlighted in dark purple colour; c) Superimposition of wild (golden yellow) and mutant (dark purple); d) Wild type p.T474 residue are shown in golden yellow colour; e) Mutant type p.T474I residue are shown in dark purple colour.
Energy Minimization and Deleterious
The energy minimization of both wild-type and mutant-type ZNF141 proteins revealed their instability, indicating a significant difference between the two variants. Deleterious analysis of the ZNF141 gene was performed using the online server Hope Server, which confirmed that the mutation occurred in a single amino acid position, specifically T474I. The mutated region was found to be larger than the corresponding region in the wild-type protein. Schematic representations of the original (left) and mutant (right) amino acids are depicted in the ZNF141 structure, with the backbone of each amino acid shown in red for the wild type and black for the mutant. These mutations are associated with severe disease annotations.
Molecular Docking
The 3D structure of ZNF141 was predicted and characterized using online computational tools. The Ramachandran plot and ERRAT indicated that approximately 90%-95% of the residues were in the allowed region [9]. The mutant-type structure was obtained from Chimera for molecular docking analysis. The ligand molecules used in the docking studies were retrieved from the ChEMBL database (CHEMBL 2106819, CHEMBL 2251733). Molecular docking analysis was used to study the binding interactions of normal and mutant ZNF141 with ligands (CHEMBL 2106819 and CHEMBL 2251733). The normal-type ZNF141 showed binding interactions with Ile220, Lys225, Phe236, Thr237 for CHEMBL 2106819 and with Lys244, Arg266, Phe267, Thr268 for CHEMBL 2251733. However, the mutant-type ZNF141 (p.T474I) showed binding interactions with different amino acids, including Lys244, His241, His245, Arg266, Phe267 and Thr268 for CHEMBL 2106819 and with Glu216, Ile220, Thr237, Thr238, His241 for CHEMBL 2251733 (Figure 4).
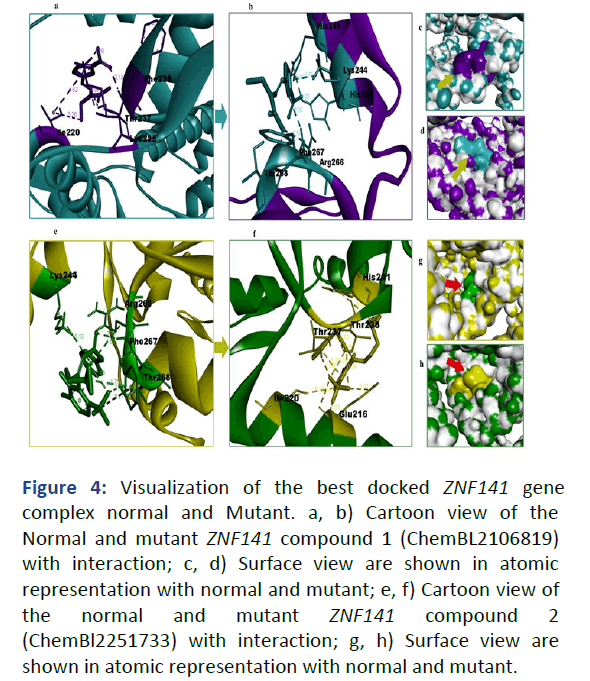
Figure 4: Visualization of the best docked ZNF141 gene complex normal and Mutant. a, b) Cartoon view of the Normal and mutant ZNF141 compound 1 (ChemBL2106819) with interaction; c, d) Surface view are shown in atomic representation with normal and mutant; e, f) Cartoon view of the normal and mutant ZNF141 compound 2 (ChemBl2251733) with interaction; g, h) Surface view are shown in atomic representation with normal and mutant.
Postaxial polydactyly type A is a congenital deformity that affects the hands or feet, characterized by the presence of an extra finger, toe or complex digital structure. The inheritance pattern of postaxial polydactyly type A is autosomal dominant and has been associated with a mutation in the ZNF141 gene located at chromosome 4p16.3 in the N-terminal region [10]. This mutation may disrupt the normal function of the ZNF141 protein, leading to abnormal limb development. The ZNF141 gene is composed of four exons that encode a protein of 474 amino acids. Analysis of the gene sequence has identified a mutation (c.1420C>T; p.Thr474Ile) that is associated with postaxial polydactyly type A. ZNF141 is a member of the zinc finger protein family and contains two C2-H2 domains, each consisting of two cysteine residues in one chain and two histidine residues in the other chain. A genetic variation in the ZNF141 gene has been identified in humans, which is responsible for causing postaxial polydactyly type A. Advancements in proteomic sequencing have provided better insights into the relationship between phenotype and genotype. Bioinformatics tools have made it easier to study this correlation by evaluating sequence variants and their potential pathogenicity. Various tools such as SNAP2, PROVEAN, PolyPhen-2, SIFT, Fathmm, predict SNP were utilized to analyze the sequence variant and determined that it is disease-causing [11].
The variant was assessed through a number of tools and was found to be deleterious. Further analysis is required to evaluate the effect of this mutation on the stability of the protein structure and its impact on the single nucleotide. Human genetic variation analysis of the ZNF141 gene was combined with computational methods to predict the potential functional impact of sequence variants and their effect on the protein's functional characteristics [12]. To evaluate the differences between the wild-type and mutanttype ZNF141 gene variants, homology modeling was performed to assess the stability of the native protein. The Root Mean Square Deviation (RMSD) value, TM score, total energy in kcal/mol of both the normal and mutated 3D models were compared. The results indicated that a single amino acid substitution resulted in a decrease in protein stability. The variant analysis and total energy calculations demonstrated that the protein was unstable, with a significant difference between the wild-type and mutant-type protein. Molecular docking studies revealed a significant conformational shift in the active binding site of the mutated ZNF141 protein, affecting its interaction with different compounds [13].
In conclusion we have reported the sequence variant of ZNF141 (c.1420C>T resulting in to single amino acid p.Thr474Ile) is predicted to be disease causing by predict SNP, SNAP2, Fathmm, I-Mutant, provean. I-Mutant predicted that there is decrease in the stability of protein structure. The variant analysis and energy calculation revealed that there is difference of kj/mol in the total energy of protein. The structure analysis of the mutant protein by chimera tool show that the threonine ring is located at the center of strand while the substituted isoleucine is hanging outside. In docking, the active binding site of mutant ZNF141 is change the stability of the protein. In results we conclude that the sequence variant (p.Thr474Ile) is changing the structural confirmation which result in decreased the stability of protein causing postaxial polydactyly typeA. In docking, the active binding site of mutant ZNF141 is change the stability of the protein. In results we conclude that the sequence variant (p.T474I) is changing the structural confirmation which result in decreased the stability of protein causing postaxial polydatyly typeA .
The authors are thankful to all members of the department of computer science and Bioinformatics, Khushal Khan Khattak university Karak, Khyber Pakhtunkhwa, Pakistan.
The authors have no conflict of interest.
S.I (Saqib Ishaq), R.T and O.H designed the study. S.I (Saqib Ishaq), R.T and O.H. performed various in silico analyses. R.T. and S.I wrote the initial MS. M.A, M.R, Z.N, A.A revised the MS. All authors have read and agreed to the published version of the manuscript.
The data that support the findings were derived from the following resources available in the public domain (GenBank) at https://www.ncbi.nlm.nih.gov/genbank/.
This study was financially supported by Higher Education Commission (HEC) Pakistan and department of computer science and bioinformatics Khushal Khan Khattak university Karak, KPK, Pakistan.
Not applicable.
Not applicable.
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
[Crossref] [Google Scholar] [PubMed]
Citation: Ishaq S, Aziz A, Habib O, Tahir R, Afzal M, et al. (2023) In silico Analysis of Human ZNF141 Gene Causing Postaxial Polydactyly Type A Disease. Res Gene Proteins. 4:12.
Copyright: © 2023 Ishaq S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction in any medium, provided the original author and source are credited.