- (2004) Volume 5, Issue 4
Naohiro Yano, LuGuang Luo
Hallett Center for Diabetes and Endocrinology, Rhode Island Hospital, Brown Medical School. Providence, RI, USA
Received: 22 December 2003 Accepted: 08 April 2004
Context Thyrotropin releasing hormone (TRH), originally identified as a hypothalamic hormone, expresses in the pancreas. The effects of TRH such as, inhibiting amylase secretion in rats through a direct effect on acinar cells, enhancing basal glucagon secretion from isolated perfused rat pancreas, and potentiating glucose-stimulated insulin secretion in perfused rat islets and insulin-secreting clonal beta-cell lines, suggest that TRH may play a role in pancreas. TRH also enlarged pancreas and increased pancreatic DNA content but deletion of TRH gene expression caused hyperglycemia in mice, suggesting that TRH may play a critical role in pancreatic development; however, the biological mechanisms of TRH in the adult pancreas remains unclear. Objectives This study explored the effect of TRH on rat pancreas. Subjects :Four male-Sprague-Dawley-rats (200-250 g) were given 10 ìg/kg BW of TRH intraperitoneally on 1st and 3rd day and sacrificed on 7th day. Four same-strain rats without TRH injection served as controls. Main outcome measures Wet pancreatic weights were measured. Pancreatic tissues were homogenized and extracted. The insulin levels of the extracts were measured by ELISA. Total RNA from the pancreases were fluorescently labeled and hybridized to microarray with 1,081 spot genes. Results TRH increased pancreatic wet weight and insulin contents. About 75% of the 1,081 genes were detected in the pancreas. TRH regulated up 99 genes and down 76 genes. The administration of TRH induced various types of gene expressions, such as G-protein coupled receptors (GPCR) and signal transduction related genes (GPCR kinase 4, transducin beta subunit 5, arrestin beta1MAPK3, MAPK5, c-Src kinase, PKCs, PI3 kinase), growth factors (PDGF-B, IGF-2, IL-18, IGF-1, IL-2, IL-6, endothelin-1) and apoptotic factors (Bcl2, BAD, Bax). Conclusion Reprogramming of transcriptome may be a way for TRH-regulation of pancreatic cellular functions.
Apoptosis; Growth Substances; Oligonucleotide Array Sequence Analysis; Pancreas; Receptors, Thyrotropin-Releasing Hormone
BAD: Bcl-xL/Bcl-2- associated death promoter; Bax: Bcl-2- associated protein X; Bcl-x: Bcl-like protein- 1; EST: expressed sequence tag; GPCR: Gproteincoupled receptor; TRH: thyrotropin releasing hormone
Thyrotropin releasing hormone (TRH) was originally identified as a hypothalamic factor with a key role in regulation of the thyroid axis [1, 2, 3, 4, 5, 6, 7]. It is generally thought that blood glucose levels can be modified by administrating TRH to the central nervous system (CNS) [8, 9, 10, 11, 12, 13, 14]. However, TRH expressed in the pancreas [15] and its actions in the pancreas include inhibition amylase secretion in rats through a direct effect on acinar cells [16], enhancing basal glucagon secretion from isolated perfused rat pancreas [17], and potentiating glucose-stimulated insulin secretion in perfused rat islets and insulin-secreting clonal beta-cell lines [18], suggesting that TRH may have direct biological effects on pancreatic function by modifying the exocrine and endocrine function of pancreas. However, the detailed mechanisms of the effects of TRH on the pancreas have not been fully elucidated.
It is well established that the structural and functional integrity of various cells is under the control of a large number of expressed genes [19]. Thus, changes in the transcription levels of several genes correlate to the state of cells. Therefore, unveiling the effect of TRH on a comprehensive profile of genes in the pancreas may facilitate understanding of the biological role of TRH in the pancreas. To achieve this objective, it is necessary to monitor simultaneously the activities of hundreds transcriptional genes. Since the mid- 1990s, cDNA arrays have been used to monitor the expressions of mRNA in multiple genes in response to various conditions and species [20, 21, 22, 23, 24, 25]. This approach has proved to be a useful tool for characterizing and quantifying the simultaneous expression of thousands of genes from specific cell populations or other tissue specimens.
To approach how TRH affects pancreatic function, we achieved TRH in vivo stimulation of pancreatic insulin productions using conservative ELISA method, and then conducted a microarray study based on the hybridization of 1,081 DNA mechanically fabricated probes. Each array was hybridized with fluorescently labeled complex targets obtained from rat pancreatic tissue with or without TRH administration. Our study suggests that this approach is not only valuable for large-scale profiling of gene transcription levels and patterns in the pancreas, but more importantly it can be used as identification of some candidate genes that may be involved in the biology of the pancreas in response to TRH. The data obtained here may serve as a resource to elucidate mechanisms of the biological role of TRH in the pancreas.
Animals
Four 8-week-old male Sprague-Dawley rats (200-250 g) were given 10 μg/kg BW of TRH intraperitoneally on the 1st and 3rd day. After 6 hours of fasting, the rats were sacrificed and pancreases were collected on 7th day. An additional four rats of the same age were injected with normal saline and served as controls. The animals were anesthetized with a single dose of intraperitoneal injection of sodium pentobarbital solution (60 mg/kg BW, Sigma, St. Louis, MO, USA). The pancreas was isolated clearly. The whole pancreas was then disconnected and moved onto ice. After being marked with only a number without knowing the treated circumstance, the pancreas was weighed by a digital balance and was snap frozen with liquid nitrogen immediately and stored at -80 °C until use for RNA preparation and for insulin assays.
Insulin ELISA
Pancreatic tissues were collected when we sacrificed the control and TRH-treated rats that we used for microarray study. Insulin concentrations in specimens were measured using Ultra Sensitive Rat Insulin ELISA Kit (Crystal Chem Inc., Downer Grove, IL, USA) according to the vender’s instruction. Briefly, insulin standards and appropriately diluted (1:50-1:500) pancreas extracts were added to insulin antibody coated 96 well microplate and incubated for 2 hours at 4 ºC. After washing 5 times, anti-rat insulin enzyme conjugate was added to the well and incubated for 30 minutes at room temperature. After washing seven times, enzyme substrate solution was added then incubated up to 45 minutes at room temperature in the dark. The reaction was stopped by the addition of 1 N sulfuric acid. Absorbance at 450 nm was read with μQuant microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA) and concentrations were calculated by KC Junior micro plate reader software (Bio-Tek Instruments Inc., Winooski, VT, USA). The experiment was repeated three times individually, giving a total sample size of 12 observations in each group.
Isolation of Total RNA
Total RNA was extracted from homogenate of frozen pancreatic tissue using TRIzol reagent (Invitrogen Life Technology, Carlsbad, CA, USA), and then treated with DNase I (Promega Corp., Madison, WI, USA) to eliminate potential contamination with residual genomic DNA then stored in ethanol at -20 ºC until use (Figure 1).
Preparation Targets and Microarray Hybridization
Four 5 μg sets of total RNA obtained from rats of the same group were mixed together. The combined 20 μg of total RNA was used for target preparation. Monofunctional cyanine fluorescent dyes (Cy3 and Cy5; Amersham Pharmacia Biotch Inc., Piscataway, NJ, USA) labeled cDNA targets were generated using Atlas Glass Fluorescent Labeling Kit (BD Bioscience Clontech, Palo Alto, CA, USA). Cy3 labeled reference target and Cy5 labeled experimental target were mixed together and hybridized to Rat Glass Microarray 1.0 (BD Bioscience Clontech, Palo Alto, CA, USA). The slides were soaked with the target solution in a hybridization chamber that provided with the array kit for 16 hours at 50 °C. After the incubation, the slides were washed for 4 times under stringent condition and then dried. Hybridized signals were scanned using microarray slide scanner (ScanArray, Perkin Elmer Lifescience, Boston, MA, USA). Strength of the signal was normalized as a ratio to the total fluorescence from all 1,081 spots as described elsewhere [26].
Microarray Analysis
To establish the comprehensive transcriptional profile of the rat pancreas, the fluorescent-labeled cDNA target generated from each specimen was hybridized to glass microarray with 1,081 genes. Identification of the expressed genes was based on a distinct position in the microarray. The gene assignment is available on Clontech web site (https://www.bdbiosciences.com/ptProduct.jsp ?backLink=ptProductList.jsp&backName=Pr oduct%20List&prodId=206131). Strength of the signal was normalized as a ratio to the total fluorescence from all 1,081 spots. In order to minimize bias arising from treatmentindependent variation in gene expression, three sets of cDNA targets were prepared from one sample and separately hybridized to the microarray. The difference in signal strength among the three hybridizations was compared gene by gene. Genes that displayed inconsistency greater than threefold normalized density validations between hybridization were excluded from the analysis.
Expressed Sequence Tag (EST) Clones
EST cDNA clones including a fragment of genes in pT7T3D-Pac vector were purchased from Invitrogen Life Technology (Carlsbad, CA, USA). Clones were seeded on ampicillincontaining LB agar plate and a single colony that grew on the plate was picked up and its insert was amplified by PCR using universal PCR primers. The sequences of the PCR primers are as follows; T7-95 upper primer, 5’-TTA ATA CGA CTC ACT ATA GGG-3’, T3-94 lower primer, 5’-AAT TAA CCC TCA ATA AAG GG-3’.
Northern Blotting
Total 10 μg RNA from each sample was run on 1% agarose gel including 1x MOPS and 2.2 M formaldehyde, then transferred to positively charged nylon membrane (Immobilon-Ny+; Millipore, Billerica, MA, USA). After transfer, RNAs were immobilized on the membrane using an automated UV crosslinker (Stratagene, La Jolla, CA, USA). PCR products from EST clones were run on agarose gel and extracted for purification. Purified PCR products were labeled with d-CTP[alpha-32P] (ICN Pharmaceutical Inc., Bryan, OH, USA) using Random Primers DNA labeling Systems (Invitrogen Life Technology, Carlsbad, CA, USA) and hybridized to the RNA membrane in 50% formamide hybridization buffer for 16 hours at 65 °C with continuous rotation. After the hybridization, the membranes were washed 4 times under high stringent condition and exposed to BioMax MS High-Sensitivity Film (Eastman Kodak Co., Rochester, NY, USA) at -80 °C for development.
Image Analysis
Images from the array hybridizations were captured and quantified using ArrayVision software (Perkin Elmer Lifescience, Boston, MA, USA). All the calculation and statistics handling were performed on Microsoft Excel (Microsoft, Seatle, WA, USA).
The pancreas tissues were harvested in compliance with an approved Rhode Island Hospital Animal Welfare Committee and Use Committee Protocol.
Statistical significance for pancreatic weight and insulin ELISA data were tested in SPSS 8.0 (SPSS Inc., Chicago, IL, USA) using the Student-t test and reported as mean±SD. Twotailed P values less than 0.05 were considered statistically significant.
Pancreatic Weight
The weight of the wet pancreas was measured immediately after dissections. TRH significantly increased the weight of the wet pancreas (1,644±116 mg, n=4) in comparison to control rats (1,394±123 mg, n=4; P=0.025) (Figure 2).
Insulin ELISA
We measured insulin levels in rat pancreatic tissue extracts to evaluate the effects of TRH on insulin synthesis. As shown in Figure 3, TRH significantly increases insulin in pancreas tissue extracts (2,412±845 ng/mg vs. control 855±115 ng/mg, normalized with protein concentrations in the extracts; n=12 in each group; P<0.001).
Figure 3. ELISA for rat pancreatic insulin content. Samples were collected simultaneously when sacrificing rats (7th day of TRH treatment). Insulin contents in pancreases were normalized with protein concentration of the extracts. Pancreas extracts from TRH-treated rats contained higher amounts of insulin than controls (2,413±845 ng/mg vs. 855±115 ng/mg, mean±SD normalized; n=12 in each group; P<0.001).
Microarray Analysis
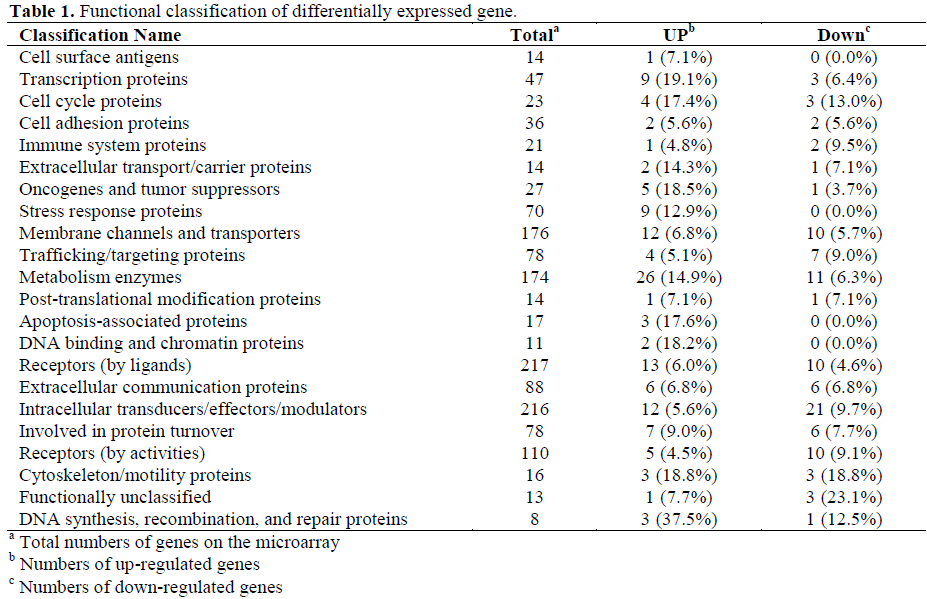
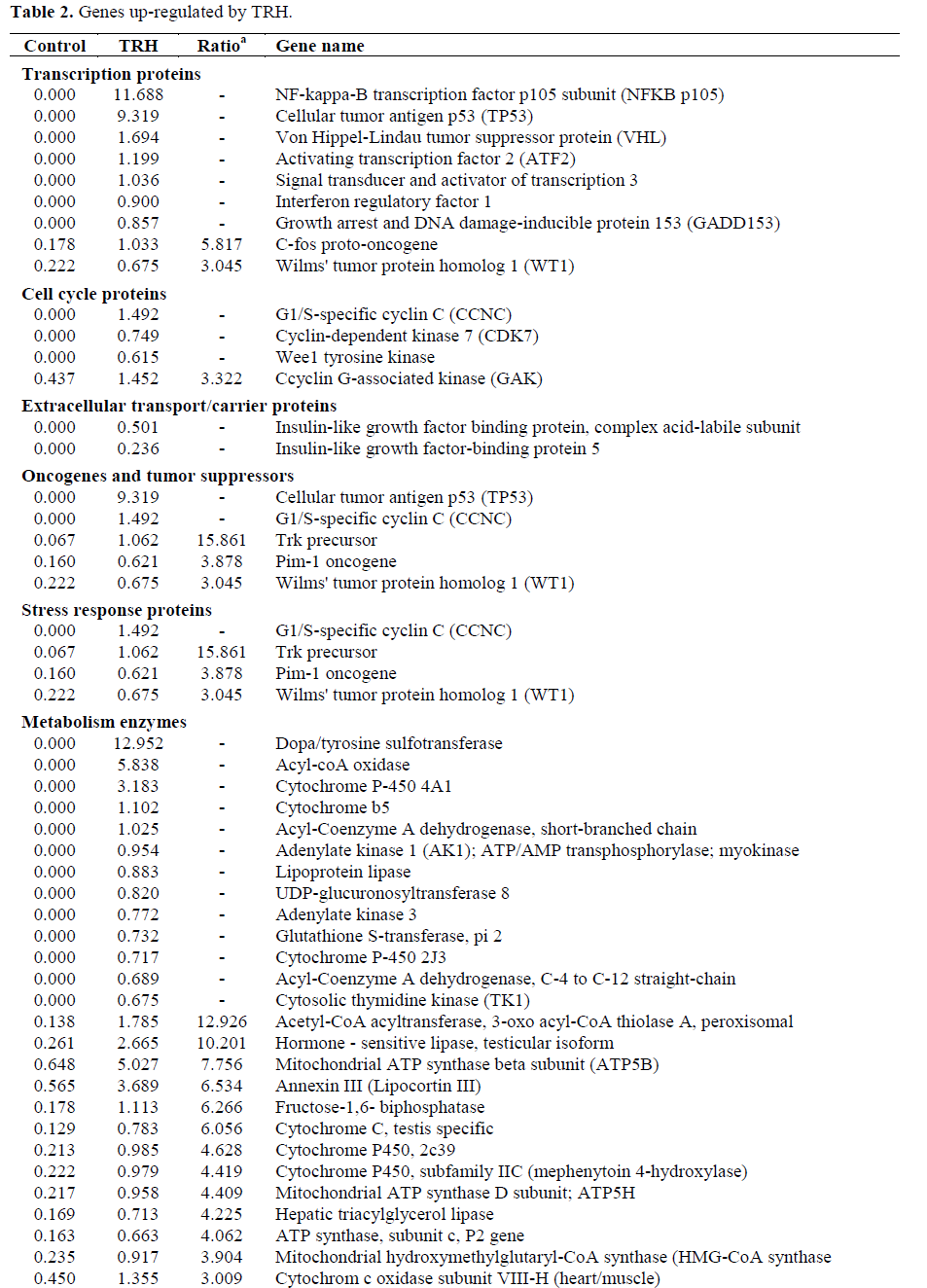
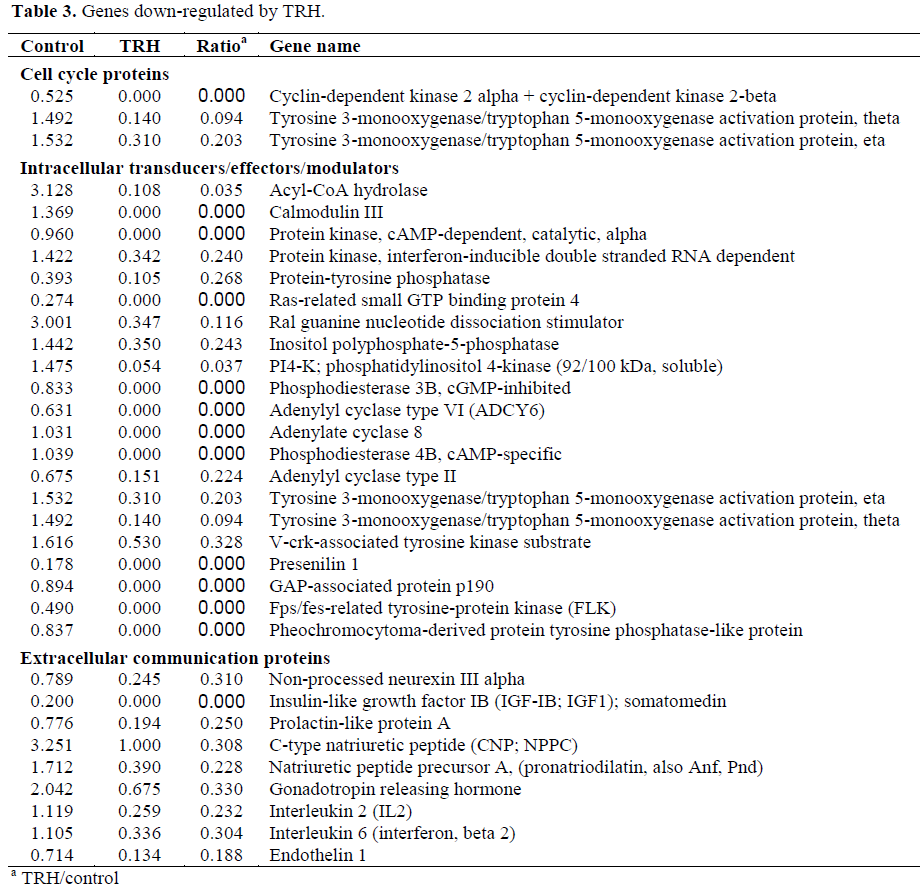
About 60-75% genes of the 1,081 genes were detected in each sample. Genes showing more than three folds difference in response to TRH treatment were counted as differentially expressed genes. The genes were classified into functions (Table 1). TRH significantly enhanced expressions of transcription proteins, oncogenes and tumor suppressors, stress response proteins, metabolism enzymes, apoptosis-associated proteins and DNA binding and chromatin proteins while suppressing immune system proteins, trafficking/targeting proteins and intracellular transducers/effectors/modulators (Tables 2 and 3).

Reconfirmation of Gene Expressions by Northern Blotting
Eight individual genes (calmodulin dependent proteinkin kinase II, arrestin-beta1, G-proteinbeta1, bcl-associated protein X, G-protein coupling receptor kinase 2, MAPK-6, n-myc, GABA-A receptor1) were reconfirmed by northern blotting. The total RNA that was used for northern analysis was taken from the same stock of RNA that we used for microarray hybridization. Probes were prepared by a random priming method using UI-EST cDNA clone as described in method section. Three screens were prepared and hybridization and stripping (using 0.4 N NaOH) were repeated four or five times. The experiments were repeated at least three times to confirm reproducibility of the results. All the 8 hybridization showed fair consistency to the results from microarray assays. Figure 4 showed representative images for all hybridizations.
Figure 4. Northern blot images for eight selected genes
from rat pancreas. Numbers under images represent
normalized density ratios of control/TRH from
microarray hybridizations. Each image included both
control left and TRH treatment right.
Identifications for the genes are as follows: A1: Gprotein
beta1 subunit (transducin); A2: arrestin beta1;
A3: calcium/calmodulin-dependent protein kinase II;
B2: G-protein coupled receptor kinase (GPK) 4; B3:
bcl-2 associated X (Bax); C1: gamma-aminobutyric
acid (GABA) A receptor; C2: N-myc; C3: MAPK3;
beta-actin (bottom row) was served as internal control.
Pancreatic beta cells possess variable neuronal derivative peptides and hormones that relate them closely with other neuronal derivatives [22, 23, 24, 25, 26, 27, 28, 29]. TRH was originally characterized and has been most extensively studied as a hypothalamic factor with a key role in the thyroid axis [1, 2, 3, 4, 5, 6, 7]. An increasing body of evidence supports the idea that the existence of TRH in the pancreas may affect pancreatic functions [30, 31, 32]. In the present study, we confirmed that TRH-treated rats had increased pancreatic mass in comparison with control rats, which was consistent with previous reports by another group [33], and improved pancreatic endocrine function by promoting beta-cell insulin production in vivo. The mechanisms of these distinct experimental findings of TRH action on the pancreas have been approached by the comprehensive pattern of gene transcription.
For the first time, a glass-based microarray with 1,081 cDNA probes has been used to characterize the profile of gene expression pattern and identify differentially expressed genes that responded to TRH in the rat pancreas. Several G-protein coupled receptor related genes (G protein-coupled receptor kinase 4, transducin beta subunit 5, Arrestin, beta1) were up-regulated by TRH. These findings suggest that effects of TRH in vivo might evoke G-protein coupled receptor (GPCR) through its cognate, consequently inducing the activation of growth relative signal transduction pathways, MAPKs (MAPK3, MAPK5) and other protein kinases (Pim-1 oncogene, c-Src kinase, protein kinase C delta, protein kinase C zeta, serine/threonine kinase 3 and inositol 1,4,5- triphosphate 3-kinase). The link between GPCR and cellular signal transduction pathways could be beta-arrestins, which are adaptor proteins that form complexes with most GPCRs following agonist binding and phosphorylation of receptors by GPKs [34]. Recent data suggests that beta-arrestins interact directly with Src family tyrosine kinases and components of the ERK1/2 and JNK3 MAP kinase modules suggest that betaarrestins may function more broadly as adapters to recruit signaling proteins for agonist-occupied GPCRs [35, 36, 37]. We previously reported involvement of Src kinase to TRH-induced trans-activation of EGF receptors in cultured beta cells [38]. The induction of beta-arrestins and c-Src may be a factor in the initiation of the pivotal role of TRH in pancreatic cell bioactivities. Upregulations of MAPK3 and other protein kinases genes’ activities that were observed in the present array study may result in arrestinrelated signal transductions leading to cell proliferations.
The proliferating effect of TRH is supposed to be synergistic with growth hormone (GH) [39], and interestingly somatoliberin, also known as growth hormone-releasing factor [40], is up regulated. The other up-regulated (PDGF-B, IGF-2, IL-18, IGF-binding protein 6) or down-regulated (IGF-1, IL-2, IL-6, endothelin-1, gonadotropin releasing hormone, pronatriodilatin, C-type natriuretic peptide, prolactin-like protein A and nonprocessed neurexin III alpha).
TRH not only stimulates pro-apoptotic gene, p53 but also enhances anti-apoptotic gene, Bcl2-like protein 1 (BclX). The results of gene array from the entire pancreas (a numerous cell types including endocrine and exocrine cells) showed anti and pro apoptotic genes simultaneously enhanced by TRH, which might indicate dual biological actions in different pancreatic cell types. Cells with predominant anti-apoptotic gene expression are subject to survival while others with proapoptotic gene expression are subject to apoptosis. The finding could lead to identification of an important mechanism in tissue repair in recovering specific functional cells while prevent over hyperplasia. Although the roles of TRH on pancreatic apoptosis have not been elucidated, this finding indicates that through enhancing antiapoptotic and pro-apoptotic factors, TRH enlargement of the pancreas could not only activate growth factor like signal transduction pathways, but also stimulate anti-apoptosis pathways. However, TRH activation of proapoptotic mechanism could be considered as a compensation for the prevention of over hyperplasia in the pancreas. The specific cell types involved in anti or pro-apoptosis activation induced by TRH requires further investigation.
Our data showed that TRH increased insulin contents in rat pancreas. Although elucidating the meaning of this increase would require more extensive studies, we found some differential expressions of genes that may be related to the synthesis or secretion of insulin. For example, Ca2+/calmodulin-dependent protein kinase (CaM kinase) was not expressed in the control rat pancreas and was induced in the TRH-treated rat. This kinase is known to be required for the activation of voltage-gated calcium channels by cAMP and the increase of Ca2+ influx into cytosol [41]. Interestingly, intracellular mobilization of Ca2+ has been disclosed to be tightly related to enhancement of insulin secretion in beta cells [42, 43, 44]. Glutamate receptor gene showed significant enhancement in expression after TRH stimulation (0.584 vs. 0.164, TRH vs. control). This receptor was reported to promote insulin secretion from beta-cell [45]. Although these data are intriguing, considering that the beta-cells occupy only a very small part of the entire pancreas, identifying insulin synthesis/secretion-related genes from the overall pancreas RNA may not be an appropriate way. Studies using isolated islets or beta-cells may elucidate this matter.
The differential gene expression pattern induced by TRH implies that the hypothalamic peptide might have the potential to be a biological regulator for pancreatic cells. Recently, peptide analogues, such as glucagons-like peptide-I, have been applied as a safe and effective candidates for recovery pancreatic beta cell loss [46, 47, 48]. However, how peptides affect gene expressions profile is still unknown and this study provided valuable information for the biological role of peptides in the pancreas.
The present study projects a certain direction to solve the long-lasting argument about the role of TRH in pancreatic functions; however, there are still many genes of which function in beta-cell biology is not fully understood. In addition, we remain concerned about some pit falls generally admitted in microarray hybridization [49].