Research Article - (2023) Volume 31, Issue 1
Received: 31-Jan-2023, Manuscript No. IPQPC-22-15543; Editor assigned: 02-Feb-2023, Pre QC No. IPQPC-22-15543 (PQ); Reviewed: 16-Feb-2023, QC No. IPQPC-22-15543; Revised: 21-Feb-2023, Manuscript No. IPQPC-22-15543 (R); Published: 28-Feb-2023, DOI: 10.36648/1479-1064.23.31.10
Background: Eukaryotic flagellum is highly conserved in basic structure and biogenesis, and defects in ciliary assembly or function lead to a wide range of human disease symptoms. The alga Dunaliella salina (D. salina), provides an excellent model for investigating flagellar/ciliary system. However, the genome it carries is unpublished. Results: To In this study, using high-throughput illumina RNA sequencing, the transcriptomes from flagella-assembling D. salina were analyzed firstly at an unprecedented depth. About 4 giga bases of raw sequence data were generated and 197,295 uni-genes were annotated with gene descriptions, conserved protein domains, or gene ontology terms against public databases. Among the annotated uni-genes, 25,412 uni-genes were differentially expressed during flagella regeneration, including 9,988 up-regulated uni-genes and 15,407 down-regulated unigenes. Moreover, to functionally categorize the D. salina uni-genes, the differentially expressed unigenes distributed into the category of biological process, molecular function and cellular component. These transcriptome datasets might reveal the mechanism of flagella assembly in D. salinacells, and serve as a public information platform for D. salina functional genomics and proteomics analysis. Furthermore, the differentially expressed unigenes involved in different signaling pathways of D. salina flagella assembly and human diseases were screened respectively.
Conclusion: These pathway-based results not only provide a further understanding to specific processes of ciliogenesis and ciliopathies, but also offer a cue to mechanism of human diseases.
Dunaliella salina; Flagella assembly; Transcriptome analysis; Functional annotation; Differential expression.
Using the domain and protein family alignment models, functional sites have been inferred from the CDD resource within the Pfam, SMART, COG, TIGRFAM and the NCBI protein clusters database. Compared against COGs, the assembled unigenes were deeply analyzed within the phylogenetically widespread domain families. According to the phylogenetic relationships, COGs were built on classifications which were grouped into 22 function categories (Figure 4). After identification of sequences, COGs consist of protein sequences encoded in 21 complete genomes, which include bacteria, eukaryotes and algae. Each COG consists of individual proteins of paralogs from at least three lineages. The 5,710 sequences showing significant homology from D. salina unigenes sets were assigned to the appropriate COG clusters. Among them, the five largest categories were “DNA recombination, replication, and repair,” “protein turnover, chaperones, posttranslational modification,” “ribosomal structure, biogenesis, translation,” “amino acid metabolism and transport,” and “general function prediction” respectively.
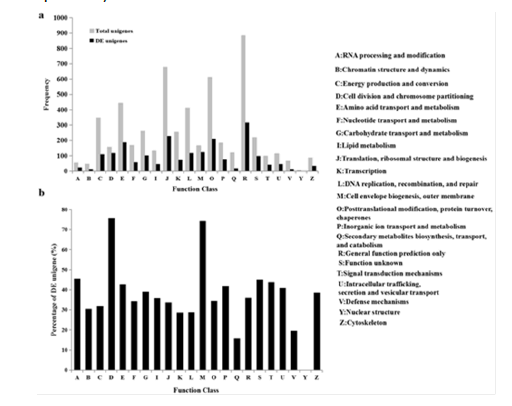
Figure 4: COG function classification of D
• The frequency of different function genes in total unigenes.
• Percentage of functional DE unigenes in total unigenes. Out of 38,678 annotated sequences, 5,710 of assembled sequences showing significant homology which had a COG classification among the 22 categories, and 1,828 differential expression (DE) unigenes among the 21 categories.
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref][Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
[Crossref] [Google Scholar] [Pubmed]
Not applicable.
All authors took part in writing, reviewing and editing the manuscript. QH Li and SY Feng designed the experiments; LQ Zhu and LN Hu performed the experiments; AF Li, SX Li and YL Li prepared the figures; Y Huang and YX Feng prepared the table. All authors reviewed the manuscript and approved it for publication.
This work was supported by the National Natural Science Foundation of China (No. U1804112), the Young Backbone Teacher Project of Henan Province Universities, China (No. 2012GGJS- 080), the Zhongjing Core Scholar’s Research Initial Fund of Henan University of Chinese Medicine (No. 00104311-2021).
The authors declare no conflicts of interest in relation to this research and its publication
Collectively, we firstly carried out a global analysis of D. salina transcriptome during flagella assembly. Using these transcriptomes datasets, the mechanism of flagella assembly in D. salina cells could be revealed, and it could be served as a public information platform for D. salina genomics and proteomics analysis. Simultaneously, other results provide a further understanding to the processes of cilium ciliogenesis or ciliopathies and a cue to the mechanism of human diseases.
So far, only 187 genes that had been implicated in 35 established ciliopathies have been identified since 1994 [14]. So, it is vital to identify more ciliopathy genes for the study of ciliary ciliogenesis and ciliopathies. Recently, many ciliogenesis proteins have been observed at extra-ciliary sites in cells [15]. Even in the absence of a primary cilium, components of ciliary machinery are repurposed for assembly and function of immunological synapse. So, D. salina transcriptome was studied by de novo assembly with purpose to identify the new functional gene. The sequencing results showed that a total of 197,295 unigenes were obtained, in which 38,678 unigenes were annotated (deposited in GenBank). Our coverage is approximately 27-fold more than all D. salina sequences deposited in GenBank (7,494 nucleotide sequences as of August 2019). In particular, the number of unigenes that hit all 6 public databases summed up to 4,139. These unigenes were assigned not only to gene or protein name descriptions, but also to putative conserved domains, gene ontology terms and metabolic pathways. However, a portion of assembled unigenes did not generate significant homology to existing genes, which could be caused by several factors, including short length of sequence, low abundance, and limitation of data base. Despite all that, the enormous data provide plenty of candidate genes for future study.
The RNA-Seq is a valuable approach for high resolution mapping of transcriptomes which is vastly superior to traditional sequencing technology. Through the detection of RNA-Seq, 22.2 and 23.3 million of 95 bp paired-end reads were generated from wild type and flagellar regenerating D. salina cells. After analysis, about 3,000 flagellar unigenes in D. salina RNA transcripts were identified, which including radial spoke protein, outer dynein arm and tubulin, flagellar motor protein kinesin family proteins and dynein, and IFT particle proteins. Among them, the majority was significantly changed, especially those in the top 111 list, containing flagella associated protein (FAP) 74, FAP117, FAP96, FAP61, FAP106, FAP63, FAP57, FAP44, IFT172, IFT81, IFT57 and IFT80. The FAP in flagella have diverse functions, such as controlling cell motility and cycle, and environment sensing [16]. The other four IFT proteins are part of complex B which interact with ciliary/flagellar precursors and transported to the flagella tip, suggesting that anterograde transport were remarkably active during D.salina flagellar regeneration [5,17]. Moreover, some post-translational modification genes were found to change greatly at mRNA level, such as HDAC and ubiquitin-protein ligase E3. These genes might play essential roles at specific steps of cilia assembly and disassembly [18,19].
Mechanism of cilium ciliogenesis or ciliopathies involves in many signaling pathways, like the Hedgehog [20] and Wnt [21]. The cilium seems to play a role in dictating the outcome of Wnt-ligand binding towards either pathway, but the details are not fully understood and data occasionally conflict. At the same time, the relationship of cilia and three important pathways in cancer, called as MAPK, mTOR and Notch signaling pathways which are revealed to involve in the during flagellar regeneration, are less reported. So, in this study, 282 KEGG pathways involved in D. salina cell development were detected for the all uingenes. As a result, GSK3β, which is involved in Wnt and Hedgehog, were significantly increased during D. salina flagellar regeneration compared that of wild type. Except that, identified unigenes were assigned to other signaling pathways, such as ErbB, P53, mTOR and Notch signaling pathways. Accumulating evidences have demonstrated that mTOR and Notch pathway has a central role not only for cell growth, but also for invasion and metastasis of tumors [22,23]. The present results showed that mTOR and Notch pathway might be active in D. salina cell during the flagellar assembly. It helped to further understand the specific processes and biological functions in D. salina flagellar regeneration, and also gave a potential reference to deep study of human diseases, like the Alzheimer’s disease, Parkinson’s disease and Huntington’s disease [6].
Using the method MARS, the differentially expressed unigenes of flagellar regenerating in D. salina cells were screened by DEGseq package. The results showed that a total of 25,412 significantly changed unigenes were detected between wild type and flagellar regenerating D. salina cells. In which, 9,988 unigenes were up-regulated and 15,424 were down-regulated, and only 10,854 unigenes were annotated with databases. Among the differential expression (DE) unigenes, 1,828 were assigned to 21 function categories without the category “Nuclear structure” (Figure 4), and the percentage of DE unigenes in total unigenes were mapped in Figure 4. The largest proportions were “cell division and chromosome partitioning” and “cell envelope biogenesis, outer membrane.” Next, the top 111 most differentially expressed unigenes were analyzed with annotation between samples (P=0), in which 82 unigenes were up-regulated and 50% of them were assigned the GO term “flagellum.” Additionally, some putative uncharacterized protein were also assigned to the GO term “flagellum” except the flagella associated proteins, such flagellar radial spoke protein, Intra-Flagellar Transport (IFT) protein, kinesin, and others. Meanwhile, some posttranslational modification kinases were differentially expressed, including methyltransferase (MetE), ubiquitin-protein ligase and S-adenosylmethionine decarboxylase. These changes might be consistent with the expectation that flagellar related proteins were changed significantly.
To search for the significantly enriched genes involved in signal transduction pathways, all the genes to terms in KEGG database were mapped during flagellar regeneration of D. salina. Compared this with wild type D. salina cells, the results demonstrated that 2,951 unigenes were differentially expressed and signaling pathway involved in flagellum were screened, including MAPK, mTOR, Wnt, Hedgehog and Notch signaling pathway, which were reported to play an important roles in cancer. Moreover, changes of genes expression during D. salina flagellar regeneration were determined by real-time PCR (Figure 7), which containing the glycogen synthase kinase 3β (GSK3β), mitogen-activated protein kinase organizer 1 (MAPK), G protein beta subunit-like (GBL), transcriptional activator LAG-3, and histone deacetylase 6 (HDAC6). The relative abundance of GSK3β, MAPK and GBL was increased and HDAC6, the key kinase to regulate flagellar resorption were decreased during flagellar regeneration. a, b, c, d, and e represented the relative abundance of GSK, GBL, HDAC, MAPK, and LAG-3 respectively. The real-time PCR results showed that GSK, GBL and MAPK were up-regulated while HDAC6 and LAG-3 were down-regulated during flagellar regeneration (*, p<0.05).
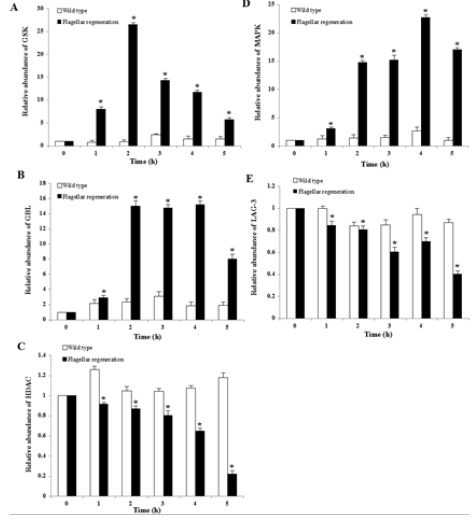
Figure 7: The changes of 5 unigenes during flagellar regeneration
The KEGG pathway database records the networks of molecular interactions in the cells, and variants of them specific to particular organisms. Compared against the KEGG database using BLASTX alignments (E-value <10-5), the assembled unigenes were annotated with corresponding enzyme commission (EC) numbers to identify the pathways activated during flagellar regeneration of D. salina. Among the 38,678 unigenes, 9,498 (24.56%) unigenes had significant matches in the database and were assigned to 282 KEGG pathways. As shown in Figure 6, the pathways most represented by unique sequences were purine metabolism (470 unigenes), spliceosome (333 unigenes), Huntington’s disease (275 unigenes), pyrimidine metabolism (256 unigenes), and RNA transport (248 unigenes) respectively. Among them, 5,282 unigenes having EC numbers were annotated to 902 enzymes. And another 417 unigenes were assigned to signaling pathway which containing cancer-related signaling pathways, like the MAPK (89 unigenes), Hedgehog (31 unigenes), Wnt (52 unigenes), ErbB (22 unigenes), P53 (21 unigenes), mTOR (19 unigenes), and Notch signaling pathways (17 unigenes). Meanwhile, Alzheimer’s disease, Parkinson’s disease and Huntington’s disease signal pathways were detected. Pathway-based analysis helped to further understand the specific processes and biological functions in D. salina flagellar regeneration.
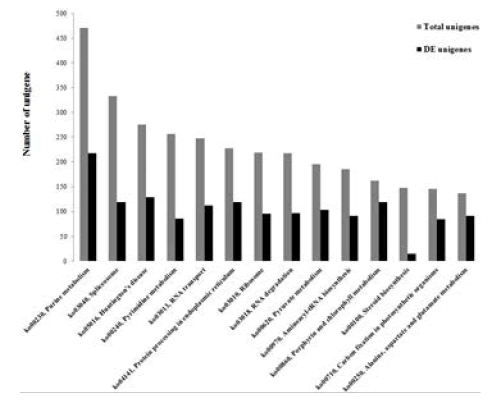
Figure 6: The histogram represented pathway assignment based on KEGG. Among the 9,497 annotated unigenes, 282 unigenes were assigned to the putative KEGG pathways, and 177 of them were affected by flagellar regeneration
Gene ontology (GO), as the dynamically-structured control vocabulary, could be used to describe functions of genes. Generally, the genes can be classified into three major categories using GO term, namely molecular function, biological process, and cellular component. So, GO term here was used to categorize each functional assembled unigenes of D. salina. Among 38,678 significant BLAST hits, 18,428 unigenes were assigned 6,384 GO terms annotations. The latter GO terms were furtherly summarized into 3 main GO categories and 44 sub-categories. Among them, 1,963 annotated unigenes were assigned the GO term “flagellar/ciliary” or “flagellum/cilium”. Molecular function, biological process, and cellular component, which are the three main GO categories, are used over in this study (Figure 5). In the cluster of biological process, most unigenes of D. salina were gathered in two sub-categories of “cellular process” (GO: 0009987) and “metabolic process” (GO: 0008152). In the cluster of cellular component, majority of unigenes were gathered in two sub-categories of “cell” (GO: 0005623) and “cell part” (GO: 0044464). At the same time, “binding functions” (GO: 0005488) and “catalytic activity” (GO: 0003824) were in the cluster of molecular function.
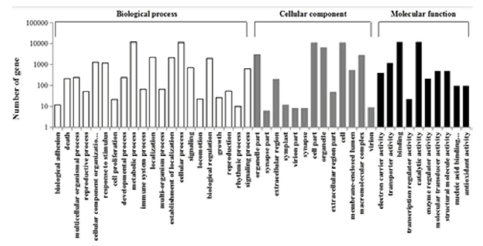
Figure 5: Gene ontology classification of the assembled unigenes
The results were summarized in 3 main categories: Biological process, cellular component and molecular function. Out of the 38,678 significant BLAST hits, 18,428 unigenes were assigned 6,384 GO term annotations, in which were further summarized into 3 main GO categories and 44 subcategories.
(D. salina) Dunaliella salina; ( PBS) phosphate-buffered saline; (ORF) open reading frame; (NR) non-redundant; (CDD) conserved domain database (COG) clusters of orthologous groups; (KEGG) kyoto encyclopedia of genes and genomes (KAAS) KEGG automatic annotation server (RPKM) reads per kilo-base per million reads; (MARS) MA-plot-based method with random sampling model; (GAPDH) glyceraldehyde-3-phosphate dehydrogenase; (SRA) Sequence Read Archive; (GO) Gene ontology; (EC) enzyme commission; (DE) differential expression; (IFT) intraflagellar transport; (MetE) methyltransferase; (GSK3β) glycogen synthase kinase 3β; (MAPK) mitogen-activated protein kinase organizer 1; (GBL) G protein beta subunit-like; (HDAC6) histone deacetylase 6; (FAP) flagella associated protein.
Based on sequence-based and domain-based alignments, D. salina transcriptome sequences and the longest sequences of contigs were annotated. Sequence-based alignments were performed against NR database from NCBI, Swiss-Prot and TrEMBL from EBI. Protein domain prediction and COG assignment were conducted by RPS-BLAST using NCBI CDD library. All the annotations from NR, Swiss-Prot, CDD, PFAM and TrEMBL database were summarized in Table 1 in which, E-value threshold was set at <10-5 and similarity >30%. However, only 38,678 unigenes had a match in the five datasets because of the relatively short length of the uni-genes (mean size of 355 bp) and the lack of D. salina genome information. Among the above-mentioned annotated unigenes, 14,660 unigenes (37.9%) were uninformative (e.g., “unknown,” “unnamed,” “putative uncharacterized” and “predicted” protein). After homologous species analysis, 18,028 of unigenes having the highest homology to genes from the Volvox carteri, followed by Chlamydomonas reinhardtii (14,603), Chlorella variabilis (3,425), Arabidopsis thaliana (3,227) and Homo sapiens (1,010). Except that, 1,087 unigenes were annonated as D. salina genes (Figure 3).
| Database | NR | SWISS-PROT | CDD | PFAM | TREMBL |
|---|---|---|---|---|---|
| Unigene number | 22,139 | 12,037 | 13,091 | 19,527 | 23,006 |
| Percentage (%) | 11.22 | 6.10 | 6.64 | 9.90 | 11.66 |
| Total unigene number | 197,295 | ||||
Table 1: Summary of the D. salina unigenes annotations
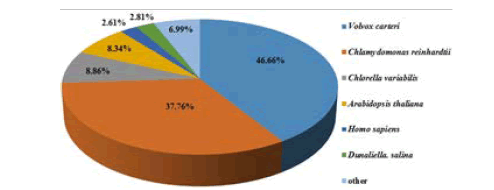
Figure 3: Results of homology search for D. salina transcriptome (E value ≤ 1.0 × 10-5)
The total transcriptome of cells treated by pH shock and untreated were separately sequenced, which generating 23,284,936 and 22,150,556 raw 95 bp paired-end reads respectively (Sequence Read Archive (SRA) submission: SUB6432910). After removal of adaptor sequences, duplication sequences, ambiguous reads and low-quality reads, 21,450,679 (96.8% of raw data) and 22,242,883 (95.5% of raw data) high-quality clean reads were obtained separately. Using de novo transcriptome assembler Velvet and Oases program, all of the clean reads were assembled and obtained 263,855 contigs which contained the untranslated regions, alternative splicing information. The results showed that the majority of contigs were between 100 bp to 500 bp, and other 10,238 contigs were greater than 1,000 bp in length. Assembly of transcriptome sequencing reads can produce more scaffolds than expressed genes, reflecting redundancy among the assembled sequences (i.e., more than one sequence per gene). Using the Gene Indices Clustering Tools, the above analyzed sequences were clustered to reduce any redundancy. Finally, a total of 197,295 “unigenes” were obtained in which 19,237 unigenes were ≥ 500 bp and 7,277 were ≥ 1,000 bp (Figure 2).
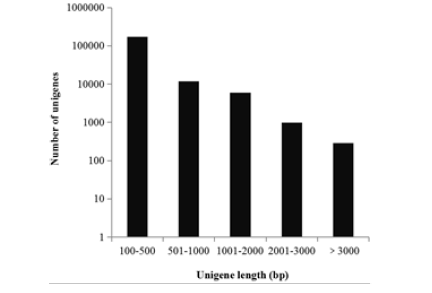
Figure 2: Assessment of unigenes length during the D. salina transcriptome assembly
To experimentally trigger the flagellar regenaration in D. salina, the flagella were removed by pH shock treatment method. As the Figure 1, the flagella were assembled to half of length at 90 min after pH shock. The growth rate of flagella reached to the maximum in the first hour. Thus, the total RNA was extracted at 1 hour after pH shock to compare the differential expression during flagellar regeneration.
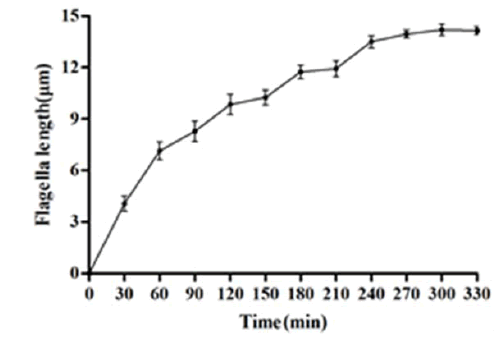
Figure 1: The regeneration curve of D. salina flagella at different time point
To further confirm the results of sequencing, 5 differential genes were compared at the mRNA level by real-time PCR. Using glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 147 bp) gene as an endogenous control for calculation of relative abundance, real-time PCR was performed using SYBR Green RT-PCR Kit (TianGen) and monitored by 7300 real-time PCR system (ABI). After that, data of relative genes expression were analyzed using the comparative CT (2-ΔΔCt) method [13].
The reads number of each unigenes was firstly transformed into the reads per kilo-base per million reads (RPKM). And then, using the method MARS (MA-plot-based method with random sampling model), the differentially expressed unigenes were identified by DEGseq package. In which, FDR ≤ 0.001 and the absolute value of log2 Ratio ≥ 1 used as the threshold to judge the significance of contig expression difference.
Using the FASTX software package, clean data from the raw data was obtained after sequencing. And then, Velvet and Oases software were used to do de novo assemble. Based on ‘Get ORF’ from EMBOSS, open reading frame (ORF) was identified by using an in-house developed program. Functional annotation of uni-genes was performed through BLAST P searches against non-redundant (NR) database from NCBI (http://www. ncbi.nlm.nih.gov), Swiss-Prot and TrEMBL from EBI (http:// www.ebi.ac.uk), followed by manual inspection. And also, the annotation of gene ontology was performed according to the homolog sequences of unigenes from these databases. In addition, using NCBI Conserved Domain Database (CDD), proteins domain prediction and clusters of orthologous groups of proteins (COG) assignment were performed by RPS-BLAST. Metabolism pathway annotation of uni-genes was performed with the kyoto encyclopedia of genes and genomes (KEGG) and KEGG automatic annotation server (KAAS) (http://www. genome.jp/kegg/).
Each frozen sample was grinded with liquid nitrogen into mortars, and then total RNA was isolated using TRIzol reagent. According to the product protocols, mRNA was further purified with MicroPoly (A) Purist Kit (Ambion, USA). Using the purified mRNA as transcription template, PCR amplification was conducted to get the double strand solexa library. Next, sequencing cluster generation and sequencing were performed on Genome Analyzer IIx (GA IIx, Illumina). The fluorescent images processing to sequences, base-calling and quality value calculation were performed by the Illumina data processing pipeline, in which 95 bp paired-end reads were obtained.
D. salina UTEX-LB-1644 was purchased from the culture collection of algae at the University of Texas at Austin, USA and cultured in modified PKS liquid medium with the optimized culture conditions. For flagella regeneration, pH shock treatment method was carried out with modified steps. Briefly, 100 mL culture of D. salina cells at logarithmic phase were collected and re-suspended in 2 ml fresh media. After that, the pH of cell suspension was quickly lowered to 2.0 by addition of 0.5 M acetic acid for 30 sec, cells were observed microscopically to confirm whether the flagella were detached. Immediately, the cells were cultured in 50 mL media with the pH value rising up to 8.0 by adding NaHCO3 within 30 s and then cultured for 6 h in culture condition. Subsequently, 5 mL cells were collected every 30 min and then fixed with 2.5% glutaraldehyde in hepes buffer (hepes 0.027 mM, NaCl 0.137 M, Na2HPO4•7H2O 0.74 mM) for several hours till the cells precipitate spontaneously. Next, the fixed cells were re-suspended in phosphate-buffered saline (PBS, pH 7.6) to coat one side of slide thoroughly to airdry. Finally, changes of flagella were observed by confocal laser scanning microscope and the length of flagella was measured. Meanwhile, before pH shock and 1 h after pH shock, cells were collected and then freezed separately for DNA extraction.
Cilium is a microtubule-based sensory organelle, which was protruded from cell surface in a variety of vertebrate cells. It acts as ‘antennae’ to sense extracellular chemical, optical signals, and developmental morphogens, such as growth factors, hormones, odorants, and transduce these signals into the cell [1,2]. And also, cilium involved in many cell process such as cell immunology and cell autophagy. Thus, defects associated with the assembly, structure, and function of cilia lead to a wide spectrum of genetic disorders collectively known as ciliopathies, including polycystic kidney disease, obesity, hydrocephalus, and even cancers [3-5].
Primary cilia are 1~10 μm length and their frequency are always disordered in ciliopathies. Abnormally low frequency or complete loss of primary cilia is commonly observed in various tumour types [6], which brings some difficulties to study their function on human diseases. Eukaryotic flagellum and cilium, essentially the same class of organelle, are highly conserved in basic structure and biogenesis spanning the eukaryotic lineage. Recently, diverse model systems such as Chlamydomonas reinhardtii, Caenorhabditis elegans, Drosophila melanogaster, and Mus musculus have been studied on the flagella/cilia. Many previous investigations have demonstrated coordination between flagellar gene expression and flagellar assembly [7]. The unicellular green alga, Dunaliella salina (D. salina ), provides an excellent model system for investigating flagellar gene expression network responses [8]. D. salina has two equal flagella with the 13 μm length, and its morphology change was easy to be caused [9]. Moreover, more genes have been found in ciliopathies and some nonciliary proteins have been reported recently [10]. Even though mitochondrial DNA, plastid DNA and nuclear genome sequence of D. salina were sequenced respectively [11,12], the proteomic or transcriptomic information of this organism is still limited, especially that of flagella. Thus, it is very necessary to identify the transcriptomic information of D. salina flagella in order to give a full complement of ciliary and ciliopathy-associated proteins.
In this study, we carried out a global analysis of D. salina transcriptome during flagella assembly. Using the illumina RNA-Seq method, more differentially expressed sequence tags during flagella assembly have been detected, which might reveal the mechanism of flagella assembly for D. salina. All transcriptome datasets will serve as a public information platform for gene expression, functional genomics and proteomics analysis in D. salina cells. In addition, some identified unigenes among the D. salina transcriptome could be used as the candidate genes to illuminate the mechanism of human diseases.
Citation: © Zhu L, Hu L, Li A, Li S, Li Y, et al. (2023) Differenial Transcriptome Analysis off Dunaliella salina during Flagellar Assembly. Qual Prim Care. 31:01.
Copyright: �© 2023 Zhu L, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.