Original Article - (2016) Volume 17, Issue 6
Gastrointestinal Oncology Service, Departments of 1Medicine, 2Pathology, 3The David Rubenstein Pancreatic Cancer Research Center, 4Surgery, and 5Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, 6Weill Cornell Medical College, New York, NY, USA
Received Date: June 02nd, 2016; Accepted Date: July 07th, 2016
Context Adenosquamous carcinoma of the pancreas represents a rare subtype (1-4%) of exocrine pancreatic cancer for which the clinical and molecular features are not well defined. Objective We reviewed clinical characteristics and performed comprehensive molecular profiling in a cohort of patients with adenosquamous carcinoma of the pancreas treated at MSKCC diagnosed from January 2000-July 2015 from a prospectively maintained database, following IRB approval to provide further insight into this disease. Design Samples were reviewed to optimally select the squamous component for molecular profiling using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets assay platform a hybridization capture based next-generation-sequencing assay for targeted deep sequencing of all exons and selected introns of 410 key cancer genes. Tumor and matched normal libraries where available were sequenced on an Illumina HiSeq 2500. Results Median overall survival for those with stage IV disease was 3.0 months (95%CI 0-13.5) and for those who underwent primary curative resection was 13.0 months (95%CI 7.5-18.5). Sixteen patients had sufficient tissue/consent for analysis. KRAS (100%), TP53 (75%) and SMAD4 (38%) alterations were most common while FAT1 (19%), JAK3 (19%), U2AF1 (6%) and PIK3R1 (6%) mutations occurred at lower frequencies. Copy number alterations such as amplification of Mcl-1(31%), MYC (6%) and FGFR1genes (6%) were detected. Conclusions Adenosquamous carcinoma of the pancreas like conventional ductal adenocarcinoma has a poor prognosis with a comparable molecular signature for the most common alterations with rarer novel alterations observed which can guide research in a disease with a need for therapeutic improvement.
Carcinoma, Adenosquamous; Pancreas; Sequence Analysis, DNA
ASCOP Adenosquamous carcinoma of the pancreas; MSKCC Memorial Sloan Kettering Cancer Center; MSK-IMPACT Memorial Sloan Kettering - Integrated Mutation Profiling of Actionable Cancer Targets; PDAC Pancreas ductal adenocarcinoma
Pancreatic ductal adenocarcinoma (PDAC) is the fourth most common cause of cancer related death with an estimated 53,070 cases occurring in the United States in 2016 [1]. The most common histological subtype of exocrine pancreas cancer is adenocarcinoma. Adenosquamous carcinoma of the pancreas (ASCOP) represents an independent entity seen in 0.4-4% of all exocrine pancreatic cancer cases with distinct clinical and histological features when compared to pancreatic ductal adenocarcinoma (PDAC) [2, 3, 4, 5]. Histologically, ASCOP is characterized by the presence of both adenomatous glandular differentiation and keratinizing squamous carcinoma constituents, with a 30% squamous carcinoma component in coexistence with ductal adenocarcinoma required to establish an ASCOP diagnosis [6, 7, 8]. The etiological development of ASCOP remains inconclusive with a number of different theories proposed [9].
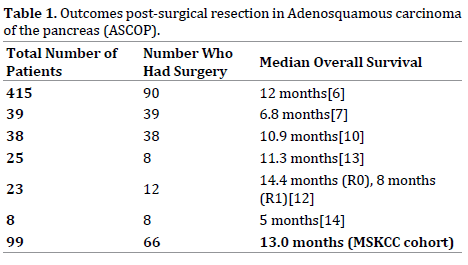
Patients with ASCOP have a clear unmet need given a poor prognosis and no defined standard treatments other than those extrapolated from PDAC. ASCOP is associated with an aggressive clinical course with poorer clinical outcomes as compared to PDAC with an increased tendency to have a larger tumor size, nodal positivity, vascular invasion or perineural invasion and be poorly differentiated [6, 7, 10, 11]. Despite surgical resection for some patients with ASCOP, survival outcomes are poor and range from 5 to 14.4 months [6, 7, 10, 12, 13, 14], see Table 1. For patients with metastatic disease, survival is dismal with a typical survival outcomes of three to four months reported [6, 11, 15].
Although progress has been in made in the molecular characterization of PDAC this has not yet translated into significant improvements in clinical outcome. Prior attempts have also been made to perform molecular profiling in ASCOP. In early reports KRAS, TP53 and SMAD4 alterations were observed [13, 16, 17, 18]. In a recent analysis, mutations in the UPF1 gene were identified in 18 out of 23 ASCOP patients [19, 20]. UPF1 encodes an RNA helicase involved in the nonsense-mediated RNA decay (NMD) pathway which selectively degrades mRNAs that harbor premature nonsense codons. Another recent study using a number of molecular techniques depicted the molecular profile of 23 patients with ASCOP identifying KRAS alterations in all patients profiled with other rarer alterations observed [21]. Recent genomic analyses of pancreas cancer have described four molecular subtypes with MYC activation, TGF-β (transforming growth factor – beta) signaling, TP53 mutations, TP63 upregulation, hypermethylation and autophagy associated with a squamous subtype [22]. The amplification of MYC has also been observed in ASCOP by whole-exome sequencing [23].
To further delineate both the clinical landscape and molecular findings in ASCOP we collected clinicopathological data on a cohort of 99 patients with ASCOP evaluated at MSKCC and used an in-house exon capture assay (Integrated Mutation Profiling of Actionable Cancer Targets, or MSK-IMPACT) to interrogate the mutation and copy number status of 410 oncogenes and tumor suppressor genes commonly altered in cancer within a subset cohort of 16 patients with available tissue [24]. In our analysis we aimed to describe the clinical outcomes for patients with this disease and identify potential actionable targets within these tumors.
Samples
Patients diagnosed with ASCOP from January 2000 to July 2015 were identified from a prospectively maintained database following IRB approval. Formalin fixed paraffin (FFPE) samples and matched blood identified for genomic sequencing were also collected under an approved IRB protocol. For all specimens, representative hematoxylin and eosin slides were reviewed by a board-certified pancreas pathologist to confirm adenosquamous diagnosis prior to submission for genomic profiling and samples were reviewed to determine the optimal component for sequencing. DNA was extracted as previously described [24].
Target Capture and Sequencing
The MSK-IMPACT assay detects mutations and copy number alterations using DNA derived from frozen and formalin-fixed, paraffin-embedded (FFPE) tissue [24]. This assay requires as little as 15 ng of input DNA and has high specificity and sensitivity using both frozen and FFPE as input. The assay has been designed to sequence all coding exons of 410 cancer-associated genes and provide 98% power to detect mutations with a true variant allele frequency of 10%, novel mutations down to a threshold of 5% variant allele frequency and mutations at recurrent hotspots down to a threshold of 2% variant allele frequency. Prior validation experiments have demonstrated the accuracy, reproducibility, and sensitivity of the MSKIMPACT assay and formed the basis of the assay approval as a clinical test by the New York State Department of Health [24]. Tumor and germline DNA from each sample was processed to generate bar-coded libraries which were subjected to exon capture using custom-designed probes. Read pairs were assigned to the corresponding tumor samples according to each barcode identity and then aligned to the reference human genome. Both tumor and matching normal DNA were run simultaneously to identify germline single nucleotide polymorphisms (SNPs). All data was then visualized and manually curated using the Integrated Genomics Viewer.
Sequence Analysis
All sequence data was processed with an extensively developed and optimized sequence analysis pipeline. Read alignment, quality, performance metrics, post-processing, somatic mutation and DNA copy number alteration detection and variant annotation were performed using standard best practices, a suite of validated open-source methods, and custom analytics. Reads were aligned to the NCBI reference human genome assembly using the Burrows Wheeler alignment tool and processed using Picard tools and the Genome Analysis Toolkit (GATK) pipeline followed by multiple sequence realignment [25, 26]. Somatic point mutations and indels were detected with the MuTect and SomaticIndelDetector algorithms, respectively, while copy number alterations were defined by seqCBS [27].
Statistics
Prespecified genomic alterations included in our MSKIMPACT assay were analyzed. For known oncogenes, we included point mutations and amplifications. For tumor suppressors, truncating mutations (nonsense, frameshift indels) and deletions were analyzed. Survival was studied using Kaplan–Meier test. Overall survival was calculated from date of diagnosis to either date of last follow up or death. Time to disease relapse post curative surgery was recorded from date of curative surgery to date of disease relapse either radiographically or biopsy proven when performed. Time to progression (TTP) on first line chemotherapy for patients with metastatic/ unresectable disease was calculated from date of initiation of chemotherapy to documented disease progression.
Ninety-nine patients with ASCOP were identified from January 2000 to July 2015, of which sixteen cases had sufficient tissue and consent for molecular analysis. Patient demographics and tumor characteristics are shown in Table 2. Median age at diagnosis was 66 years (21- 88) with a male predominance (60.6%). The majority of patients had stage II (54.5%) disease at diagnosis followed by stage IV (28.3%); stage III (6.1%) and stage I (4.0%). Tumors had location preponderance to the head of the pancreas (56.6%). Sixty six patients underwent curative resection of which 12 (18%) remain without evidence of disease after a median follow up of 22 months (1-132). Of the 66 patients who had curative intent resection, thirty-nine (59.1%) had positive lymph nodes at time of surgical resection. The median recurrence free interval post curative surgical resection was four months (1-51). Forty-three patients had disease recurrence post surgery with the liver the most commonly affected site in 74.4% of cases, followed by lung (13.9%), peritoneal (11.6%) and local only recurrence in 4.7%.

We reviewed adjuvant chemotherapy regimens utilized in ASCOP. Forty patients of 66 resected; (61%) received adjuvant therapy in our institution. With regards to other patients; nine patients followed up locally, nine (14%) patients had metastatic disease on first radiographic imaging post surgery, three patients did not receive therapy, two patients died postoperatively, two received neoadjuvant therapy and one patient had a prolonged hospital stay post surgery which precluded adjuvant therapy. The most frequent adjuvant regimen was gemcitabine based therapy in twenty two patients. Fluropyrimidine based therapy was used in eight patients. The combination of gemcitabine and fluoropyrimidine (capecitabine in all cases) was employed in a separate 8 patients and two patients enrolled to a clinical trial. The addition of a platinum agent to gemcitabine or fluropyrimidine based regimen occurred in four cases (4/40; 10%). Sixteen patients (40%) received chemoradiation as part of their adjuvant therapy regimen.
Six patients had locally advanced stage III disease. One patient achieved surgical resection following neoadjuvant GTX (gemcitabine, docetaxel and capecitabine) followed by capecitabine based chemoradiation. Pathology revealed a ypT2N0M0 tumor, this patient developed a solitary metastatic lung nodule 22 months post surgery which was surgically resected. The patient remains without evidence of disease on follow up; 34 months from diagnoses and 5 months post lung metastasectomy.
Median overall survival (OS) for resected patients in our cohort was 13 months (95%CI 7.5-18.5), Figure 1. The median OS for patients with stage IV disease was 6 months (95%CI 0-13.2) Figure 2. With regards to first line metastatic systemic chemotherapy regimens; gemcitabine based therapy was utilized in 28 patients, fluropyrimidine based in 17 patients, gemcitabine plus fluropyrimidine based therapy in 4 patients and other regimens in two patients [platinum plus irinotecan (n=1) and docetaxel (n=1)]. Of these patients, 22 (45%) received a platinum as part of therapy. Median TTP on first line chemotherapy for patients with metastatic/unresectable disease was three months (range 1-11). Six patients with metastatic disease were treated with best supportive care alone, three had de novo metastatic disease and three patients with disease relapse. Two patients with metastatic disease declined chemotherapy.
Genomic Alterations Seen in ASCOP by MSK-IMPACT
With the aim of assessing somatic genetic alterations in ASCOP, we analyzed 16 patients with ASCOP who had both available tissue and consent using a capture-based, massively parallel, next-generation sequencing assay (MSK-IMPACT). Of the 16 patients, 10 had available matched normal blood for comparison with the tumor sample. Of the 16 patient samples sequenced, 14 were from the primary tumor site and two samples were from metastases (liver; n=2), Table 3. This likely represents a sampling bias given ASCOP diagnosis may be challenging from FNA sampling as compared to primary surgical specimens. The majority of somatic alterations seen were single base substitutions including non synonymous coding changes, nonsense mutations with the remainder amplifications or deletions.

MSK-IMPACT identified a variety of somatic alterations, Figure 3a. KRAS mutations were seen in all 16 patient samples (100%) similar to previous reports [14, 21, 28]. Fifteen of the KRAS alterations were at codon 12 with one at codon 61 (Q61H). TP53 mutations were seen in 12/16 (75%) of samples. Molecular alterations within SMAD4 (38%) and CDKN2A (38%) occurred along with a variety of other copy number alterations, Figure 3b. Within the MSK-IMPACT set of 16 patients, two presented with stage IV disease and fourteen had stage II disease at initial presentation (stage IIA; n=5, stage IIB; n=9), Figure 3a,b. Twelve of the fourteen patients who had primary resection developed metastatic disease with alterations in KRAS (100%; 12/12), TP53 (83%; 10/12), SMAD4 (42%, 5/12), CDKN2A (42%, 5/12) most commonly identified in these patients. All five patients with SMAD4 alterations (missense; n=2, truncating mutations; n=3) detected by MSK-IMPACT in the primary tumor post resection developed metastatic disease with a median time to disease relapse of 7.5 months (range 5-16) as well as all patients with Mcl-1 amplification (n=5) who had a median time to disease relapse of 6 months, (range 1-16). One other patient with a SMAD4 truncating mutation had stage IV disease at presentation, Figure 3a.
The Phosphoinositide 3-Kinase (PI3K) pathway was altered in 25% samples (4/16) through alterations in PIK3R1, PIK3R2, PIK3CA and PTEN loss. We identified a PIK3R1 missense E683K mutation in one patient sample while the PIK3R2 I556 missense mutation has not been previously characterized. The PIK3CA mutation was a known oncogenic helical domain E542K missense mutation. Mutations in the FAT1 gene located on 4q35.2 were seen in 3 (19%) patient samples. FAT1 missense mutations observed included an R2567H missense mutation (n=1), a T3424M missense mutation (n=1) and both an M2845I and V2582M missense mutation in one patient. The R2567H mutation has been previously reported in a lung adenocarcinoma [29]. Two patients had TGFBR1 (Transforming Growth Factor, Beta Receptor 1) in frame mutations resulting in TGFBR1 loss.
STK11 (LKB1) is a tumor suppressor gene for which one of its targets is the tuberin protein which inhibits mTOR1 [30]. One patient had an STK11 missense mutation, F354L. This mutation was previously described in the germline of a child who developed neuroblastoma and a follicular variant of papillary thyroid cancer [31]. It also was observed in the germline in one Finnish patient and as a somatic mutation in Korean patients with left sided colon cancer [32, 33]. In a previous study in patients with left sided colon cancer the F354L variant was seen not to alter LKB1 kinase activity and felt to have a non-cancer inducing effect [33]. Two TSC1 missense mutations were identified; one T360N missense mutation previously described and putatively linked to autism spectrum disorder and felt not to be pathogenic in the tuberous sclerosis database [34]. The functional significance of the TSC1 V407M is unknown. One patient had an RNF43 truncation mutation.
We also identified some original alterations. One patient had an FGFR1 amplification, not been previously described in ASCOP. One patient had a U2AF1 S34F missense mutation. U2AF1 mutations have been noted in cancers such as AML, squamous head and neck carcinoma, endometrial carcinoma, urothelial bladder cancer and breast, lung and colon adenocarcinoma previously [35]. U2AF1 alterations occur in ~11% of myelodysplastic syndromes causing aberrant gene splicing with the S34F missense mutation the most frequent mutation [36]. This mutation to our knowledge is the first time U2AF1 has been described in ASCOP. 31% (5/16) of patients had Mcl- 1 (Myeloid cell leukemia-1) amplification. Mcl-1part of the anti-apoptotic subclass of Bcl-2 proteins and modulated by oncogenic pathways such as MAPK, mTOR and PI3K signaling has been described in pancreas adenocarcinoma but to our knowledge not in ASCOP [37, 38, 39, 40].
Adenosquamous cancer of the pancreas is a relatively rare but particularly virulent form of pancreas cancer. From our analyses and other previous reports ASCOP is a systemic disease underpinned by poor survival outcomes. Despite surgical resection with curative intent a median OS of 13 months is very poor even relative to PDAC [6, 7, 10, 12, 13, 14] (Table 1). The rate of lymph node positivity at time of surgery was high (59.1%), similar to previously reported rates of 51.4% [6] and 76% [10]. In our cohort of 66 patients who had resection, nine (13.6%) had evidence of metastases at time of first radiographic assessment postoperatively. Relapse free interval post surgical resection in our cohort was short at 4 months (1-51) with disease most commonly affecting the liver (74.4%). These factors suggest that a neoadjuvant systemic therapy approach may be more appropriate in ASCOP thereby selecting a more appropriate patient population to undergo future major resection.
Currently, no standard therapy exists for ASCOP in the neoadjuvant, adjuvant or metastatic setting. Adjuvant chemotherapy in ASCOP has been associated with an improved survival in some single institution nonrandomized series [13, 41]. In contrast neither adjuvant chemotherapy nor radiation improved outcome in another analysis [5]. Given the lack of established therapy most regimens are based on those used in PDAC. No survival difference was seen between gemcitabine based, 5-FU based or combination of gemcitabine and 5-FU adjuvant therapy in a prior retrospective analysis [41]. In our sample set, gemcitabine based therapy was most frequently employed in the adjuvant setting, with a doublet of gemcitabine and capecitabine employed in eight separate patients and platinum doublet in four patients. The use of platinum agents in the adjuvant setting has been associated with an improved survival of 19.1 months as compared to 10.7 months in a non-platinum group (p=0.015) in a single institution non-randomized retrospective analysis with no difference in outcome observed between cisplatin or oxaliplatin [41]. In our cohort, 10% of cases received adjuvant platinum therapy (oxaliplatin n=1, cisplatin n=3) in combination with fluropyrimidine or gemcitabine based therapy. Notably, none of our patients sequenced displayed mutations in BRCA1, BRCA2, or PALB2 genes that otherwise infer platinum sensitivity. In a prior report, adjuvant chemoradiation (CRT) provided a survival benefit as compared to surgery alone with characteristics such as; tumor size of ≥3 cm, presence of vascular or perineural invasion, nodal metastases and a squamous component ≥30% benefiting from CRT [10]. Incorporation of CRT as part of the adjuvant treatment regimen in our cohort of patients post resection was 40%. Given the small numbers (16 patients) and retrospective nature of our data definitive conclusions are not possible. Prospective trials with multi institutional collaboration are required to definitely corroborate the role of platinum and radiotherapy in the adjuvant setting.
For those with metastatic disease prognosis was dismal with a median OS of 6 months (95%CI 0-13.2) observed. Gemcitabine based therapy was most frequent, with a platinum agent utilized in 45% of patients who received systemic chemotherapy. This rate was higher than our 10% platinum rate in the adjuvant setting. Nevertheless, median time to progression (TTP) was three months (1- 11) underscoring the inherent need to develop improved therapeutic strategies for this aggressive disease. Putatively this may be possible by understanding the molecular and genomic drivers of this disease with the hope to develop novel therapeutic agents.
MSK-IMPACT results for the 16 ASCOP tumors sequenced revealed alterations in TP53, KRAS, SMAD4 and CDKN2A, similar to those found in PDAC. KRAS mutations were seen in 100% of our samples which is similar to previous reports [14, 16, 21]. Given the high prevalence of KRAS mutations this highlights the need to develop improved strategies to identify the Achilles heal of mutated RAS. We did identify a number of potentially actionable targets as well as novel alterations which may represent susceptibility for therapeutic manipulation. 25% of patients had PI3K pathway alterations. Single agent PIK3CA inhibition has minimal single agent activity in pancreas cancer and given the cooccurrence of KRAS in all cases, single agent PIK3CA inhibition is likely to have a negligible therapeutic effect. Therefore combination strategies with either chemotherapy or MEK inhibitors ostensibly are needed. Of note, Weiss et al described an ASCOP patient with both a KRAS and PIK3CA alteration identified by next generation sequencing and a combination of a PI3K inhibitor and MEK inhibitor was commenced with a transient response [42]. In addition PIK3R1 mutations may be a potential target and are under assessment in endometrial cancer [43]. Other potential amenable targets include CDKN2A loss which can be potentially targeted with CDK4/6 or MDM2 inhibitors [44]. One patient had RNF43 loss which in vitro is sensitive to porcupine inhibitors [45]. Novel alterations in JAK3 were seen (19%) with in vitro activity observed with the JAK3 inhibitor CP-690550 in ALL with the V722I mutation, the same mutation identified in one of our samples [46]. FAT1 mutations were observed in 19% of cases and are linked with functional consequences upon Wnt pathway dysregulation and activation [47]. Additionally, one FGFR1 amplification not previously described in ASCOP may be amenable to FGFR inhibitor therapy. Mcl-1 altered in 31% of cases has been tested in vivo and in vitro in pancreas cancer and are targets which may benefit a subset of patients [48]. This could be an important target since all five patients with Mcl-1 amplification by MSK-IMPACT in their primary tumor developed metastatic disease post resection.
As previously stated genomic analyses has characterized a squamous type within four molecular subtypes of pancreas cancer with characteristic signaling networks including TP53 mutations and MYC activation [22]. By MSK-IMPACT, 75% had TP53 mutations and one patient had MYC amplification. MYC amplification is associated with a poor outcome in ASCOP [23]. Our patient with MYC amplification developed metastatic disease four months post surgery and had a survival time of four months from development of metastatic disease. Two patients both never smokers with a head of pancreas primary location had TGFBR1 in frame mutations resulting in TGFBR1 loss both, an alteration described in other squamous cancers of the head and neck and cervix [49, 50]. One patient had a KDM6A mutation in our cohort which along with TP53 has been associated with a pancreas squamous subtype [22]. In comparison to another previous study of molecular profiling in ASCOP our rates of TP53, SMAD4, PIK3CA, and EGFR alterations were higher with identical KRAS (100%), BRAF (0%) and NRAS (0%) frequencies [21]. In addition, we noted that all patients with SMAD4 alterations and Mcl- 1 amplification developed metastatic disease post surgery, although numbers are small both SMAD4 and Mcl-1 have previously been implicated as possible prognostic factors [51, 52].
The etiology of ASCOP development is uncertain [9]. Our molecular analysis identified a GNAS alteration in one patient in a known hotspot (R201H) occurring with a KRAS mutation suggesting a highly MAPK dysregulated tumor. GNAS alterations are associated with tumors that arise from a precursor intraductal pancreatic neoplasm (IPMN) and are felt to be specific for this event [53]. The presence of a GNAS mutation in this case may infer an IPMN precursor lesion, a rare event which has been previously described [54]. In addition adenosquamous carcinoma has also been described to occur in a mucinous cystadenoma [17].
In conclusion, our sequencing platform identified a molecular signature for frequent alterations within commonly affected PDAC genes (KRAS, TP53, SMAD4 and CDKN2A); however novel genes in U2AF1, PIK3R1 and copy number alterations within Mcl-1 and FGFR1 were identified. ASCOP is associated with poor outcomes despite curative resection with a high rate of nodal positivity, short interval to disease relapse and poor survival despite surgery observed. Furthermore outcomes for metastatic disease are underwhelming, therefore ASCOP represents a disease with a clear need for improvement. Future efforts can hopefully bridge the gap between molecular complexity and therapeutic actionability for ASCOP.
We gratefully acknowledge funding from the Herman Bott Pancreatic Cancer Research Fund and the Suzanne Cohn Simon Pancreatic Cancer Research Fund in support of this project.
The authors have no potential conflict of interest.