- (2015) Volume 16, Issue 3
Vy Nguyen, Scott Hurton, Subhashini Ayloo, Michele Molinari
Department of Surgery, Dalhousie University, Halifax, Nova Scotia, Canada
Received January 20th, 2015- Accepted April 07th, 2015
Metabolomics is an emerging scientific field focusing on compounds, also known as metabolomes that are produced by biologic systems. Metabolomes represent phenotypic end-products of living entities, and are of particular interest as they represent a novel group of biomarkers that can be used for diagnosis, monitoring response to treatments, and more recently as potential therapeutic agents for benign but more so for malignant diseases. One of the main factors associated with improved survival of patients affected by malignancies is early diagnosis, particularly for those tumors with poor prognosis such as pancreatic cancer. Currently, there are no diagnostic tests for the screening of pancreatic cancer, and although novel biomarkers continue to be discovered, none has yet to be proven useful for this purpose. The main aims of this paper are to review the current literature and to summarize the most relevant advances in the field of metabolomics applied to pancreatic adenocarcinoma.
Early Diagnosis; Early Detection of Cancer; Metabolomics; Pancreatic Neoplasms
Metabolomics is an emerging branch of studies focusing on phenotypic characteristics rather than genetic profiles of biological systems [1, 2]. Metabolomes are defined as all compounds generated by living organisms during their life cycles and reflect the function of the organisms as a whole [1]. This concept can be applied to single cells, organisms or larger biologic systems. Because of the dynamic profile of living entities, the intricate interactions of biochemical cycles and the uniqueness of each system, metabolomics is one of the most challenging disciplines of the family of “omics” [2].
Contrary to genomes that remain relatively stable over time, metabolomes are subject to environmental influences that can alter their profile in a very short period of time and the mere sampling, preparation and analysis of biological samples may alter their outlines [2]. This has proven to be one of the hardest tasks to overcome for the advancement of this field. Another challenge is the variability of the molecular size and the large number of metabolites within a given sample that requires very sophisticated methods for the extraction and for the data analysis [2]. Despite these challenges, the field of metabolomics has made significant advances, and in recent years, has become one of the most promising areas of research for some malignancies where pure genetic tests might fall short especially when exogenous predisposing factors play a role [3].
Metabolomic analysis can be applied to both in vitro and in vivo specimens by using body fluids, cells or tissues. Various methods have been utilized to detect and generate metabolic profiles. The two most common are nuclear magnetic resonance spectropscopy (NMR) and mass spectrometry (MS) [2-4]. These can be coupled with gas chromatography, liquid chromatography or capillary electrophoresis [2-4]. NMR has the benefit of requiring minimal sample preparation; it can analyze a wide range of metabolites and applies to both solids and liquids. On the other hand, MS has the benefit of being more sensitive than NMR but does require more extensive sample preparation that might lead to interactions between analysates and instruments [3, 4].
Metabolomics in Oncology
The use of metabolomics seems to have the greatest potential in oncological research, as the pathogenesis of cancer often is due to both genetic and environmental factors. Comparing variations of metabolic profiles occurring in neoplastic cells versus normal cells, can lead to the identification of various metabolites or combinations of metabolites that could potentially be used for the diagnosis of specific cancers or to monitor their response to treatments.
In addition, metabolomics can be used to analyze a large number of compounds and biological samples that can be used for for screening and early detection. This includes the use of a wide variety of methods such as obtaining tissues or using less invasive methods such as collecting biofluids like serum, saliva or urine [2-4]. Another benefit of metabolomics is the lower sample volume of no more than 0.5 ml for sufficient analysis.
In recent years, the study of metabolomics has expanded to other fields of research such as pharmacometabolomics where metabolomes are analyzed before, during and after treatment allowing researchers to better understand how patients respond to specific drugs and how neoplastic processes react to established or experimental therapies [5].
Metabolomics and Pancreatic Cancer
Pancreatic adenocarcinoma (PC) is the twelfth most common cancer in North America and Europe, and the fourth cause of cancer related deaths with an overall 5-year survival rate of 5 to 10% [6-9]. The high mortality rate of PC is multifactorial and when PC is diagnosed in early stage and patients undergo resection, 5-year survival can be as high as 20-35% [9]. However, the majority of patients become symptomatic late and are diagnosed with locally advanced or with metastatic disease.
Presently, no screening tests have shown any promising impact for the survival of PC patients [10]. Cross sectional imaging tests such as abdominal ultrasound (US), computerized tomography (CT-scan), magnetic resonance imaging (MRI), endoscopic ultrasound (EUS) and endoscopic retrograde cholangiopancreatography (ERCP) have failed to detect early stage tumors when used for screening purposes even in high risk populations [10-12]. Serologic markers such as carbohydrate antigen 19-9 (CA 19-9) lack enough specificity to differentiate between PC and other gastrointestinal malignancies or benign pancreaticobiliary diseases such as pancreatitis and cholangitis [10]. Newer serologic biomarkers (CA242, M2-PK, PBF-4, PNA-binding glycoprotein, hTert, MMP-2, synuclein-γ, ICAM-1, OPG, CEA, TIMP-1, MUC1, CEACAM1, MIC1) have been identified as potential screening tools but none has been proven to be better than CA 19-9 [10, 13-23].
Applications of Metabolomics in Pancreatic Cancer
One of the main barriers to early detection of PC is differentiating it from benign pancreatic pathology most notably chronic pancreatitis. Numerous research groups have sought to differentiate between the two conditions using NMR and MS. NMR provides a broader approach for the identification of various metabolites whereas MS provides the ability to identify individual metabolite markers [3]. Each technique has individually been applied in the field of early detection of PC (Table 1).
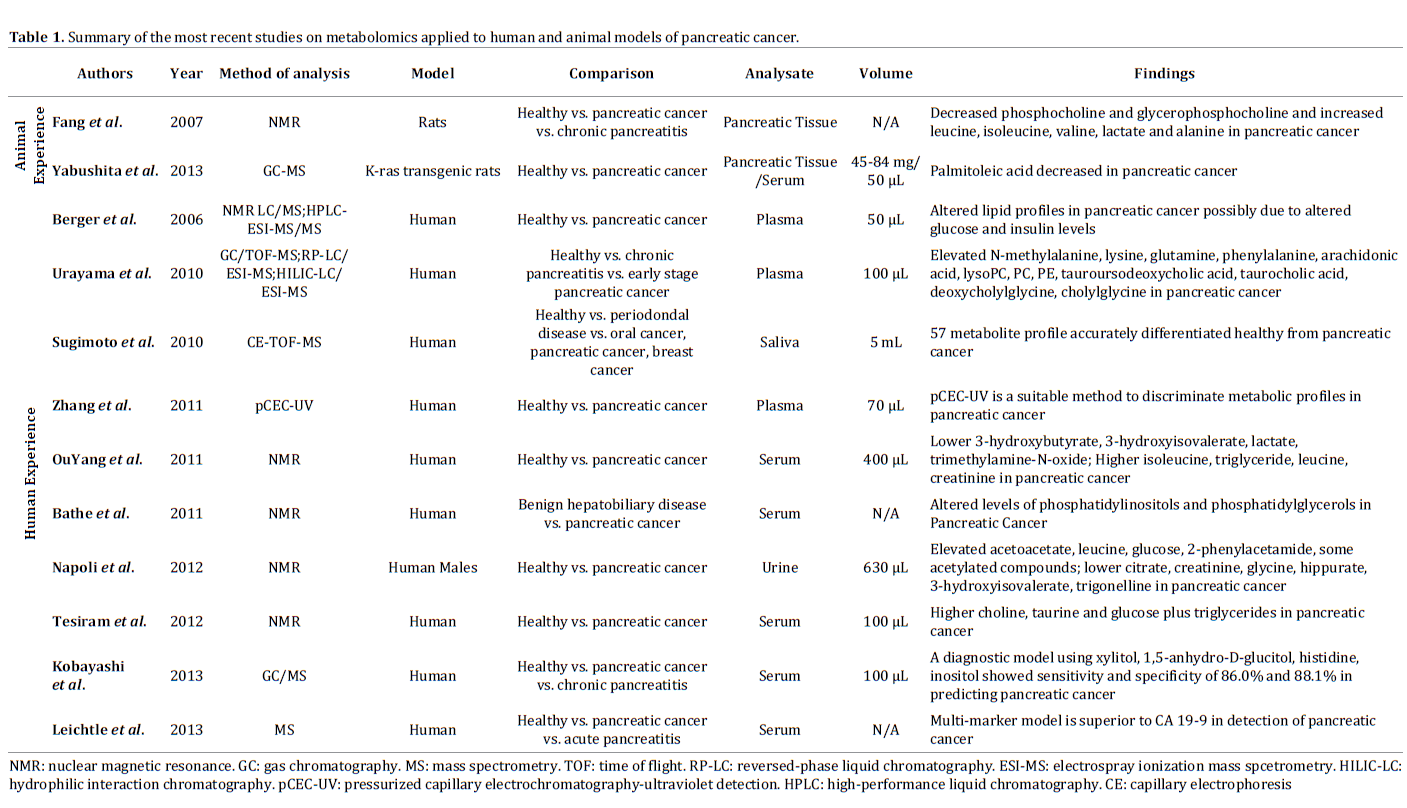
Animal Experience
Animal studies have applied metabolomics mainly to rats for early diagnosis of PC. Yabushita et al. used K-ras 301 transgenic rats to compare metaobolic profiles of PC vs. controls. This rat model had previously shown to be a valuable model to study the development of PC once rats are treated with an adenoviral vector [24]. In their study, rats were confirmed to have PC without evidence of local invasion or metastases. The authors found that rats with PC had decreased serum levels of palmitoleic acid when compared to healthy controls on gas chromatographymass spectroscopic analysis [25]. Fang et al. also used rat models that were divided in three different groups: rats with PC, rats with chronic pancreatitis and healthy controls. PC rats developed moderately differentiated PCs and demonstrated differences in their metabolic profiles in comparison to rats with chronic pancreatitis and healthy controls as phosphocholine and glycerophosphocholine were decreased while leucine, isoleucine, valine, lactate and alanine were increased. The opposite trend was seen in chronic rats with pancreatitis [26].
Human Experience
Several research groups have applied metabolomics techniques to patients with PC and compared their profiles to healthy controls (Table 1). NMR technologies allowed Tesiram et al. to identify statistically significant different levels of choline, taurine glucose and triglycerides between the two groups. Studies were performed on patients using 100 μL of serum and tumor clinical stages varied from IIB to IV [27]. NMR was also used by Ou Yang et al. and reported altered levels of isoleucine, triglycerides, leucine, creatinine, lactate, 3-hydroxybutyrate, 3-hydroxyisovalerate, and trimethylamine-N-oxide. In this study, only 400 μL of serum was necessary [28]. Urayama et al. used multiple methods including gas chromatography/ time-of-flight mass spectrometry (GC/TOF-MS), reversedphase liquid chromatography/electrospray ionization mass spectrometry (RP-LC/ESI-MS), and hydrophilic interaction chromatography/electrospray ionization mass spectrometry (HILIC-LC/ESI-MS) to identify 24 metabolites that were abnormal in patients with PC. These patients had early stage PC (IB-IIB) and only 100 μL of serum was used [29]. Zhang et al. were able to demonstrate a novel method of pressurized capillary electrochromatography/ultraviolet detection (pCEC-UV) as a superior method than established chromatographic techniques for PC [30]. Berger et al. utilized 1D proton NMR techniques as well as liquid chromatography (LC/ MS), and high performance liquid chromatography (HPLCESI- MS/MS) to demonstrate the alterations of various lipid molecules in PC patients with tumor stages between I to IV where only 50 μL of plasma was needed [31]. These studies have shown that over time, techniques and volumes used for their analysis have changed, but no single metabolome has consistently shown to predict the presence of PC with enough accuracy. Therefore, further studies are necessary to figure out what analytical methods perform better in these settings.
Because no single molecule has shown to be promising enough for the discrimination of patients with early PC, Kobayashi et al. have used multiple logistic regression analysis to compare PC patients with healthy controls. PC patients varied from stage 0 to IV and 100 μL of plasma was used. Using GC/MS, 18 metabolites were identified as potential biomarkers and using 4 of these metabolites (xylitol, 1,5-anhydro-D-glucitol, histidine, and inositol), a statistical diagnostic model was generated. This model performed extremely well with an area under the receiveroperating characteristic curve (AUC) of 0.92. When compared to conventional tumor markers CA19-9 and CEA in the validation study, this model did not perform as well as the AUC went down to 0.76 but maintained acceptable sensitivity (71.4%) and specificity (78.1%). The sensitivity of this diagnostic model remained overall stable even when applied to early stage diseases (0-IIB) (77.8%) in comparison to CA 19-9 (55.6%) and CEA (44.4%) [32].
Differentiating Between Benign and Malignant Disease
Bathe et al. [33] used NMR spectroscopy to identify serum metabolites that could be used to differentiate various benign versus malignant pancreatic diseases. In their study, glutamate, acetone, and 3-hydroxybutyrate were strongly associated with malignancy and their multivariate model demonstrated good discrimination with an AUC of 0.83 [33]. The robustness of this model was further demonstrated via a feasibility test of 14 pairs matched for the presence of jaundice, pancreatic mass and diabetes with comparable AUC values. Similarly, Leichtle et al. [34] performed a three-class analysis using serum from individuals affected by PC (stages I-IV), pancreatitis and healthy controls. A significant difference among the three groups was found for 22 amino acids [34]. Using mass spectrometry both individually and in conjunction with CA19-9 levels, a multivariate model based on both specific amino acids in conjunction with CA19-9 provided a 3-Dimensional analogue of AUC (VUS) with good discrimination (VUS value=0.89) [34]. These studies have shown that metabolomics, both alone and in conjunction with other established serologic testing, can be used to improve our current ability to detect and discriminate PC from other benign conditions.
Early Detection of Pancreatic Cancer
Beyond serum, other biofluids have been used to study metabolic profiles for the diagnosis of early PC [35, 36]. Previous investigations of bile and pancreatic fluids had identified a range of proteins that could be used as markers [10]. Unfortunately these samples require invasive techniques that are costly and often not available in many centers. To overcome these limitations, several studies have focused on the use of biofluids that can be collected by noninvasive techniques. For example, 5 mL of unstimulated saliva obtained from patients with non-metastatic PC was analyzed using capillary electrophoresis time-of-flight mass spectrometry (CE/TOF-MS) and compared with samples from healthy controls, patients with oral or breast cancers and periodontal diseases. Using multiple linear regression analysis of a total of 48 potential metabolites, 5 were selected to construct a model that showed excellent accuracy for the detection of PC with AUC equal to 0.94, the highest value ever seen [35] for any of the diagnostic tests. Furthermore, Napoli et al. [36] performed a case controlled study using NMR spectrography on urine samples (630 μL) from PC patients (stages II-IV) and healthy individuals. Not only they observed a statistically significant different profile between the two groups, but they were also able to identify differences between different cancer stages [36]. This was the first study that was able to discriminate diverse metabolomic profiles for patients with different stages of PC. Although extremely promising, further studies are necessary to confirm if these results are reproducible in other populations.
Future Directions and Applications of Metabolomics in Pancreatic Cancer
The studies reviewed in this paper have all sought to identify useful bio-markers that could, one day, be used for the diagnosis of early PC either by improving the sensitivity and specificity of current tests or substitute them. So far, metabolomics have shown promising results but not sufficient to change current clinical practice and more studies are needed [33, 34]. Given the fact that metabolomic profiles incorporate both genetic and environmental factors that play a role in PC, it is important that future studies identify and control for some of these variables (e.g. age, sex, co-morbidities, and disease stage).
Another promising area where metabolomics can have a significant impact is the analysis of samples from groups with pre-malignant or predisposing factors for PC such as intraductal papillary mucynous neoplasms, familial PC, BRCA2 and other conditions.
The concept of pharmacometabolomics was introduced earlier in this review. Although pharmacometabolomics have not been widely used in clinical practice, expanding its approach to include changes following resection and/or chemoradiation therapy in PC would allow metabolomics not only to be applied to diagnosis and screening but also to surveillance.
Although metabolomics applied to PC is still in its infancy, it has the all the potentials of becoming an important field that could lead to the identification of better biomarkers to be used alone or in combination with other already available modalities for the screening, early detection, staging and treatment of this challenging tumor. Hopefully, this would translate in better prognosis for patients with PC whose survival continues to be unsatisfactory.
The authors had no conflicts of interest