Keywords
Cl-/HCO3 —ATPase; GABAAR subunits; Phosphorylation; Rat brain; Proteomics
Abbreviations
GABAAR: GABAA receptor; EtOH: Ethanol; PMP: Plasma membrane-enriched preparation; PEP: Purified enzyme preparation; hrCNEPAGE: High resolution clear native Electrophoresis; SDS-PAGE: Sodium dodecyl sulfate polyacrylamide gel electrophoresisIntroduction
Introduction
The neuronal Cl-/HCO3--ATPase (Cl--transporting ATPase; EC 3.6.3.11) is a "molecular machine" involved in Cl--transport across the membrane, driven by ATP consumption [1]. The Cl-/HCO3- -ATPase is of interest primarily because this protein functions both as a transport ATPase and as a receptor [2]. It is classified as a P-type transport ATPase based on the formation of a highenergy phosphorylated (acyl phosphate) intermediate during ATP hydrolysis, its sensitivity to phosphorylation inhibitors and SH-reagents, and its specificity for nucleotides and anions [2]. Nevertheless, the catalytic subunit of the protein complex remains to be identified. However, allosteric regulation of this enzyme function by classical activators and blockers of the GABAA receptors of the GABAA receptors (GABAARs) indicate a proximity of the receptor binding sites and the ATP-hydrolysis centre on the protein molecule [2,3]. In addition, preliminary results on the purified protein have indicated the presence of [3H] muscimolbinding subunits in a purified enzyme preparation, suggesting a structural coupling to the inhibitor receptors [3].
GABAA receptor/Cl--channels are the major inhibitory receptors in brain postsynaptic membranes and they interact with γ-aminobutyric acid (GABA) to induce the transport of Cl- (and/or HCO3-) by an electrochemical gradient, causing hyperpolarization or depolarization of the membrane potential [4,5]. Nineteen subunits (α1-6, β1-3, γ1-3, δ, ε, θ, π, and ρ1-3) are presently recognized that can occur in various combinations to form distinct GABAAR structures [6]. However, the most common receptor ensembles have a composition of one γ, two α, and two β-subunits [5-7]. All subunits may be involved in the formation of the ion pore (i.e., the Cl--channel), but only the 3 subunits have the unique properties of anion selectivity [8], and these subunits alone possess the ability to form an ion pore [9]. Studies on the ATPase indicate a selective activation by different anions and especially by both Cl- and HCO3- [1,2]. These findings have indicated the importance of investigating the presence of GABAAR β-3 subunits in the purified protein complex.
The available literature shows that one of the major factors regulating the function of the ion-transporting membrane systems is phosphorylation [10,11]. Various studies have confirmed that the GABAAR β1-3 subunits are the primary substrates for phosphorylation by different protein kinases [12] and were therefore assumed to exist in ATPases that have bound inhibitory receptors [13]. A number of studies have reported the existence of GABAAR ensembles that are directly phosphorylated by ATP without an involvement of protein kinases [14,15]. In particular, electrophysiological experiments on rabbit vestibular neuron membranes have shown the presence of a "molecular machine" with properties of the GABAARs [15]. These "atypical" GABAAR structures are suggested to serve as devices that extrude Cl- after postsynaptic GABA interaction, at the expense of ATP consumed in their phosphorylation. However, this type of GABAAR has not been investigated at the molecular level. Identification of ATP-dependent and receptor-coupled Cl--transport systems is therefore an important step in determining the molecular mechanisms responsible for the processes of the inhibition/ excitation of the central nervous system.
One strategy for further identification of the enzyme would be to include the use of methodological approaches that would allow the demonstration of GABAAR peptide subunits within a purified enzyme preparation. Some of the available literature has shown that 2-DE gel electrophoresis BN-PAGE/SDS-PAGE allows the separation and identification of various membrane-associated protein complexes, including enzymes and receptors [16,17]. In addition, application of gel-based proteomics techniques involving western blot analysis (and/or mass spectrometry) can also aid in the analysis of site-specific phosphorylation or other post-translational modifications [17].
With these possibilities in mind, we devised the following strategy for characterization of the Cl-/HCO3--ATPase protein using gelbased proteomic techniques and western blot analysis. We first detected the test protein in gels strips after 1-DE Clear Native- PAGE based on Pi staining after preincubation with Mg2+-ATP in the presence or absence of anions and GABAAergic blockers (e.g., EtOH, picrotoxin). The GABAAR subunits were then detected by western blot analysis using antibodies against GABAAR β1-3 subunits. This was followed by a further determination of the peptide subunits constituting the oligomeric protein complex by 2-D hrCNE/SDS-PAGE gel electrophoresis and autoradiography. Direct phosphorylation of the protein subunits, without the involvement of protein kinases, was confirmed by pre-incubation with γ-32 P-ATP and Mg2+.
Experimental Procedure
Chemicals
Gamma-aminobutyric acid (GABA), picrotoxin, Tris(hydroxymethyl)aminomethane, (4-(2-hydroxyethyl)- 1 - p i p e r a z i n e e t h a n e s u l f o n i c a c i d ) ( H e p e s ) , N a 2 AT P, ethylenediaminetetraacetic acid (EDTA), Triton X-100, sodium dodecyl sulfate (SDS) BioUltra, Protease Inhibitor Cocktail Tablets (Roche Applied Science), Kit for High-Molecular Weights were obtained from Sigma-Aldrich (USA). 6-Aminohexanoic acid ≥98.5% (Fluka). Electrophoresis reagent kit (Bio-Rad Laboratories, USA). Coomassie Blue G-250 (Serva Blue G) was purchased from Serva. Resin Toyopearl AF-Epoxy-650 M (Tosoh Bioscience, Japan). Low-molecular mass markers were obtained from Invitrogen (Thermo Fisher scientific, USA). [γ-32P]ATP (5×10–6 dpm/nmol) (Amersham, Biosciences, United States Biochemical Corp.). The other chemicals were the analytical grade and locally obtained.
Animals
Male Wistar rats (∼240 g) were obtained from vivarium of the Federal State Scientific Institution “Research Institute of General Pathology and Pathophysiology”. Animals were housed in a climate-controlled room on a 12–12-hr light/dark cycle and free access to water and food. The experimental investigations of the material was approved by the Ethical Committee of FSBSI “Institute of general pathology and pathophysiology (No 01- 01/147 from October 12, 2009) and performed according to the principles expressed in the Declaration of Helsinki revised by WMA, Fortaleza, Brazil, 2013.
Total brain membrane preparation
All procedures were performed at 0°C to 4°C. After decapitation of animals, the brain was isolated, homogenized in 8 vol. of ice-cold buffer solution containing 0.25M sucrose, 1 mM ethylenediaminetetraacetic acid-Tris (hydroxymethyl) aminomethane (EDTA-Tris, pH 7.4), 25 mM N-(2-Hydroxyethyl) piperazine-N-(2-ethanesulfonic acid) (HEPES-Tris, pH 7.4), protease inhibitor cocktail tablets (Roche Applied Science), and centrifuged in a Beckman ultracentrifuge (SW-28 bucket rotor) at 10000 g and 4°C for 25 min. The supernatant was centrifuged at 50000 g and 4°C for 1 hr. The supernatant was discarded and microsomal fraction-enriched plasma membranes (pellets) were resuspended in 1 mM EDTA-Tris (pH 7.4), 25 mM HEPES-Tris (pH 7.4), stirred for 15 min, and centrifuged (50000 g, 1 hr). The resulting pellets were resuspended in 25 mM HEPES-Tris (pH 7.4).
Sucrose gradient centrifugation for plasma membrane enrichment
The plasma membrane purification procedures were carried out as described previously, followed by sucrose density gradient ultracentrifugation [17]. Each gradient, starting from the bottom of the tube, was composed of 4 ml each of 69%, 35%, 20%, 15% and 10% w/v sucrose solution.
Total membrane preparations were layered on top of the sucrose cushion. The sample was centrifuged at 4°C at 70000 g for 3.5 hr. After centrifugation, fractions from 10% to 20% sucrose interface were collected and diluted ten times with homogenization buffer and then centrifuged at 4°C at 100000 g for 30 min. After discarding the supernatant the pellet was stored at 80°C until use. This plasma membrane-enriched preparation (PMP) was used for further ATPase purification and measurements of the enzyme activity. Protein concentration was estimated according to Bradford protocol using bovine serum albumin as the standard [18].
hrCNE-PAGE
High resolution Clear Native Electrophoresis (hrCNE-PAGE) was carried out according to a previously published protocol [16]. The PMP and the PEP were suspended in membrane protein extraction buffer containing 500 mM 6-aminocaproic acid, protease inhibitor cocktail tablets (Roche Applied Science), 60 mM imidazol/HCl, 1 mM EDTA, pH 7.0. After resuspension, a 20% Triton X-100 stock solution was added at a ratio of 1:10 to achieve a final 2% Triton X-100 concentration. Membrane protein extraction was performed for 1 h at 4°C with vortexing with followed centrifugation for 1 h at 1,20,000 x g, at 4°C. The supernatant for hrCNE (around 2 ml) was supplemented with 200 μl of 50% glycerol, 500 mM 6-aminocaproic acid and 0.1% Ponceau S. Equal sample volumes (30 μl) were applied to multiple 0.5 × 0.15 cm gel wells. The actual protein load for each lane (around 80 μg of protein). The hrCNE-PAGE gel buffer contained 500 mM 6-aminocaproic acid, 75 mM imidazole/HCl, pH 7.0; the anode buffer (25 mM imidazole/HCl, pH 7.0; cathode buffer (50 mM Tricine, nonionic detergent (0.05% Triton X-100 and 0.01% DDM), 7.5 mM imidazole, pH 7.0) were used for hrCNE. hrCNEPAGE was performed in VE-20 (Helikon, Russia) with 20×20 cm2 glasses, 1.5 mm thickness spacer using a 3.5% stacking and a 4-15% separating gel. For electrophoresis, the voltage was set to 75 V for 1 h, 110V for 5 hr, and was increased sequentially to 250V (maximum current 15 mA/gel), until the dye front reached the bottom of the gel. hrCNE-PAGE wards (a 300 kDa band marked) representing together the ATPase complex and GABAAR subunits β3 were cut out and gel put in a falcon tube with buffer and mildly shake in the cold overnight. After, the evolution solution with protein was concentrated and dialysis to be used for hrCNE/SDSPAGE (2-DE). High-molecular mass markers were obtained from Invitrogen (Thermo Fisher scientific, USA).
SDS-PAGE
The fractions enriched with enzyme activity were boiled for 5 min in an SDS treatment buffer consisting of 62.5 mM Tris, 10% glycerol, 5% 2-mercaptoethanol, 4% SDS, and 0.001% bromophenol blue [19]. Samples (~15 μg/protein well) were applied to 12% SDS-PAGE according to Laemmli at 30 mA current. Electrophoregrams were stained with Colloidal Coomassie Brilliant Blue (G-250) or detected by autoradiography of ATP-γ- 32P binding. A vertical electrophoresis chamber VE-20 (Helikon, 270 Russia) with 20 × 20 cm2 glasses, 1.5 mm thickness spacer. The power supply was a Power Pac HC (250 V/3 A/300 W; BioRad). Electrophoresis was carried out at C with an initial current of 75 V (during the first hour). Then voltage was increased to 130 V for the next 7 hr, and increased to 170 V until the dye front reached the bottom of the gel. Low-molecular mass markers, PageRuler were obtained from Invitrogen (Thermo Fisher scientific, USA).
Affinity chromatography
The affinity purification of the enzyme was carried out as described previously [3]. The resin for affinity chromatography was prepared by coupling muscimol (Sigma, Aldrich) to Toyopearl AF-Epoxy-650M (Tosoh Bioscience, Japan). Briefly, the soluble enrichment plasma membranes fraction (15 ml) was applied to the column (2.5 × 5 cm2) in 25 mM HEPES-Tris (pH 7.4), 0.2% Triton X-100 and washed at 45 ml/hr with 500 ml of 25 mM HEPES-Tris (pH 7.4) containing 0.1 mM EGTA, 1 mM MgCl2, and 0.2% Triton X-100. The enzyme was specifically eluted 30 ml/ hr with 30 ml HEPES-Tris (pH 7.4) containing 5 mM GABA, 0.2% Triton X-100, 0.1 mM EGTA. Fractions (0.5 ml) were collected. Prior to Native- and SDS-PAGE, each sample was concentrated to 0.5 ml using centrifugal concentrators Vivaspin Turbo 15 (100 kDa) and Vivaspin Turbo 4 (100 kDa), (Manufacturer Browser, Sartorius Vivascience). The concentrated sample was used as purified enzyme preparation (PEP) [3].
ATP hydrolysis assay
Detection of the Cl-/HCO3 Pase activity was carried out as published previously [3]. Briefly, the enzyme preparation (~7 μg) was added to 0.5 ml incubation medium containing 25 mM HEPES-Tris buffer (pH 7.4), 3.0 mM MgSO4, 3.0 mM ATP-Tris, 5 mM NaCl/25 mM NaHCO3 and 60 mM NaNO3 (neutral salt) to measure enzyme activity. The specific ATPases activities were estimated from the increase in the content of inorganic phosphorus (Pi) in 0.5 ml incubation medium at 30C for 20 min. Phosphorus concentration in samples (0.25 ml) was measured by of the modification method of Hess and Derr [20]. Membrane samples were preincubated at 30C for 20 min with the relevant GABAAergic ligands in incubation medium containing 25 mM HEPES-Tris buffer (pH 7.4), 5 mM NaCl/25 mM NaHCO3 and 60 mM NaNO3. The reaction was started by addition of the substrate (Mg2+-ATP) to the incubation medium. The Cl-/HCO3--ATPase activity were determined in the absence and in the presence of Cl-/HCO3-.
In gel catalytic activity assays
The in gel assays followed the principles described by Witting et al. with the following modifications [16]. Cl Pase complex was optionally assayed in consecutive steps using the same gel strip. To quantify the ATP hydrolysis activity of the Cl-/HCO3- Pase complex, two similar gel strips from the same native gel were preincubated in parallel for 1.5 hr in 25 mM HEPES–Tris (pH 7.4) at 40°C containing or not containing the anions or GABAAergic ligands. Following removal of the incubation solution, the gels were incubated in assay buffer (25 mM HEPES–Tris (pH 7.4, 5 mM MgSO4, 5 mM ATP, 0.2% Pb(NO3)2, containing or not containing chemicals. ATP hydrolysis correlated with the development of white lead phosphate precipitates. Moreover, gel strips for Pi staining carried similar with protocol as measured the ATPase activity, also [20]. Here, substrate hydrolysis correlated with the development of green phosphate precipitates. The reaction was stopped using 50% methanol for 30 min, and then the gel was transferred to water and scanned using a Bio-Rad scanner.
Western blot test
In Western blots the proteins in gel strips after hrCNE (1-DE) and subsequent hrCNE/SDS-(2-DE) were transferred to a PVDF membrane at 17 mA for 60 min were transferred onto PVDF membrane. After blocking with 10% non-fat dry milk in 0.1% TBST membranes for 1 h at room temperature and then incubated overnight at 4°C with primary antibodies diluted as followed: anti-GABAAR β3 antibody (1:5000) (ab98968, Abcam, Cambridge, United Kingdom), anti-GABAAR β2 antibody (1:10000) (ab15600, Abcam, Cambridge, United Kingdom) and anti-GABAAR β 1 antibody (1:1000) (ab154822, Abcam, Cambridge, United Kingdom). On the next day, after washing 3 times with 0.05% Tween-20 and phosphate buffered saline, the membrane was incubated for 1 hr at room temperature with the corresponding horseradish peroxidase (HRP). conjugated secondary antibodies, anti-rabbit IgG (Cell Signaling Technologies) according to the supplier’s protocol. The electro blotting apparatus and power supply were the Trans-Blot SD semi-dry electrophoretic transfer cell and Power Pac HC (250 V, 3A, 300 W), respectively (both from BioRad).
Protein phosphorylation
Purification by affinity chromatography the Cl-/HCO3--ATPase was reconstituted into proteoliposomes as described before [1]. The proteoliposomes was phosphorylated in 50 μl of incubation medium containing 25 mM MOPS–Tris (pH 6.0), 3 mM MgSO4, and protein (approximately 45 μg). The reaction of phosphorylation was started by the addition to the incubation medium of 70 μM ATP-γ-32P (specific radioactivity, 5 × 10–6 dpm/ nmol) (Amersham, Biosciences). The mixture was incubated at 0°C to 1°C for 2 min. To study the effect of 5mMCl-/25 mM HCO3-, NH2OH (50 mM) or EtOH (500 mM) on the phosphoprotein formation, the membrane preparation was preincubated with the ligands at 0°C to 1°C for 20 min. Proteoliposomes with purified enzymes preparation were applied to 12% SDS-PAGE (MOPS Denaturing Running Buffer, pH 6.0) and detected by autoradiography of 3 2P. The enzyme preparations were boiled for 5 minutes in a SDS treatment buffer consisting of 50 mM MOPSTris (pH 6.0), 10% glycerol, 5% 2-mercaptoethanol, 2% SDS, and 0.001% bromophenol blue. Stained and dried gels were placed in a chamber for autoradiography (Sigma, USA) on a Hyperfilm™ MP film (Amersham, USA) and exposed at room temperature for 96 hr. The film was developed using the standard developer to obtain the maximum contrast image.
Results
Cl-/HCO3--ATPase and GABAAR detection
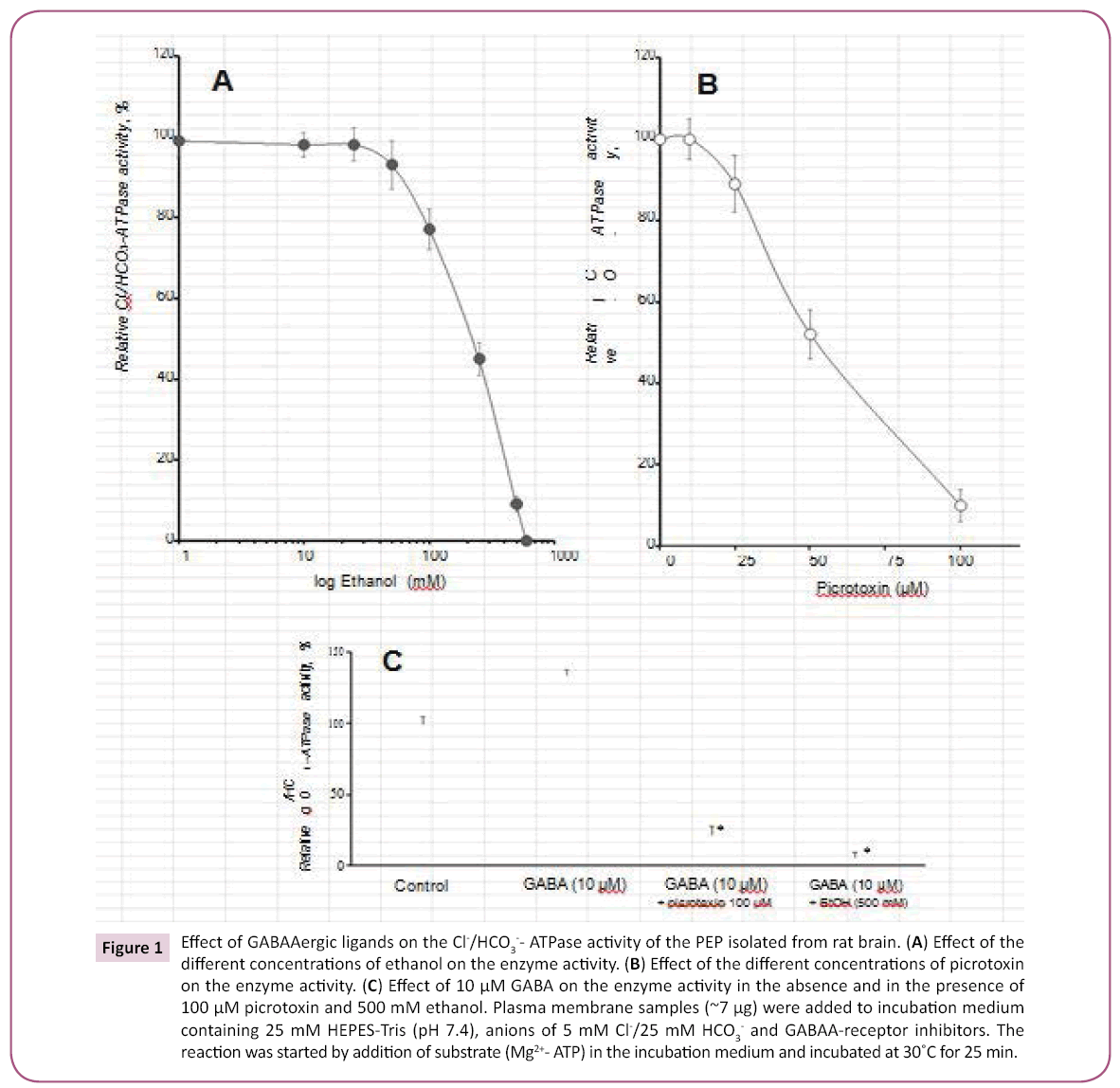
Previously, we have shown that rat brain plasma membranes contain an ATPase that is sensitive to GABAAergic ligands and that this ATPase is activated in the presence of 5 mM Cl-/ 25 mM HCO3- (Cl-/HCO3--ATPase) [21]. The activity of the isolated and affinity-purified enzyme preparation prepared in the present study was 230.0 μmol Pi/h/mg protein and we used this activity as 100% (Figure 1A). Investigation of the effect of a number of GABAAergic ligands (EtOH, picrotoxin and GABA) on the enzyme activity, as shown in Figure 1A,B, revealed that EtOH (200-500 mM) and picrotoxin (30-100 μM) completely inhibited the Cl-/ HCO3--ATPase activity.
Figure 1: Effect of GABAAergic ligands on the Cl-/HCO3-- ATPase activity of the PEP isolated from rat brain. (A) Effect of the different concentrations of ethanol on the enzyme activity. (B) Effect of the different concentrations of picrotoxin on the enzyme activity. (C) Effect of 10 μM GABA on the enzyme activity in the absence and in the presence of 100 μM picrotoxin and 500 mM ethanol. Plasma membrane samples (~7 μg) were added to incubation medium containing 25 mM HEPES-Tris (pH 7.4), anions of 5 mM Cl-/25 mM HCO3- and GABAA-receptor inhibitors. The reaction was started by addition of substrate (Mg2+- ATP) in the incubation medium and incubated at 30˚C for 25 min.
Both inhibitors - picrotoxin (100 μM) and EtOH (500 mM) also eliminated the activating effect of 10 μM GABA on the enzyme activity (Figure 1C).
The selective sensitivity of the Cl-/HCO3--ATPase to the tested GABAAergic ligands allowed the detection of the enzyme in gel strips after 1-D hrCNE-PAGE. A native-PAGE gel revealed that the PMP yields about ten protein bands, whereas the PEP was resolved as a single protein band with a molecular mass of 300 kDa (Figure 2A). The gel strips for both the PMP and the PEP preparations revealed ATPase activity after preincubation with Mg2+-ATP (Figure 2B). The ATPase activity in the PMP, unlike the PEP, was observed in several bands with molecular weights of 300-900 kDa. We confirmed the identification of the protein with the molecular weight 300 kDa as the purified ATPase by preincubating the gel strips in an incubation medium containing 5 mM Mg2+-ATP and 5 mM Cl-/25 mM HCO3-, in the presence and in the absence of picrotoxin (100 μM) or EtOH (500 mM). As shown in (Figure 2C), the addition of Cl-/HCO3- to the incubation medium increased the intensity of the Pi staining in the gels. Addition of picrotoxin to the incubation medium significantly inhibited the ATPase activity observed in the gel strips, while the addition of ethanol completely eliminated the ATPase activity.
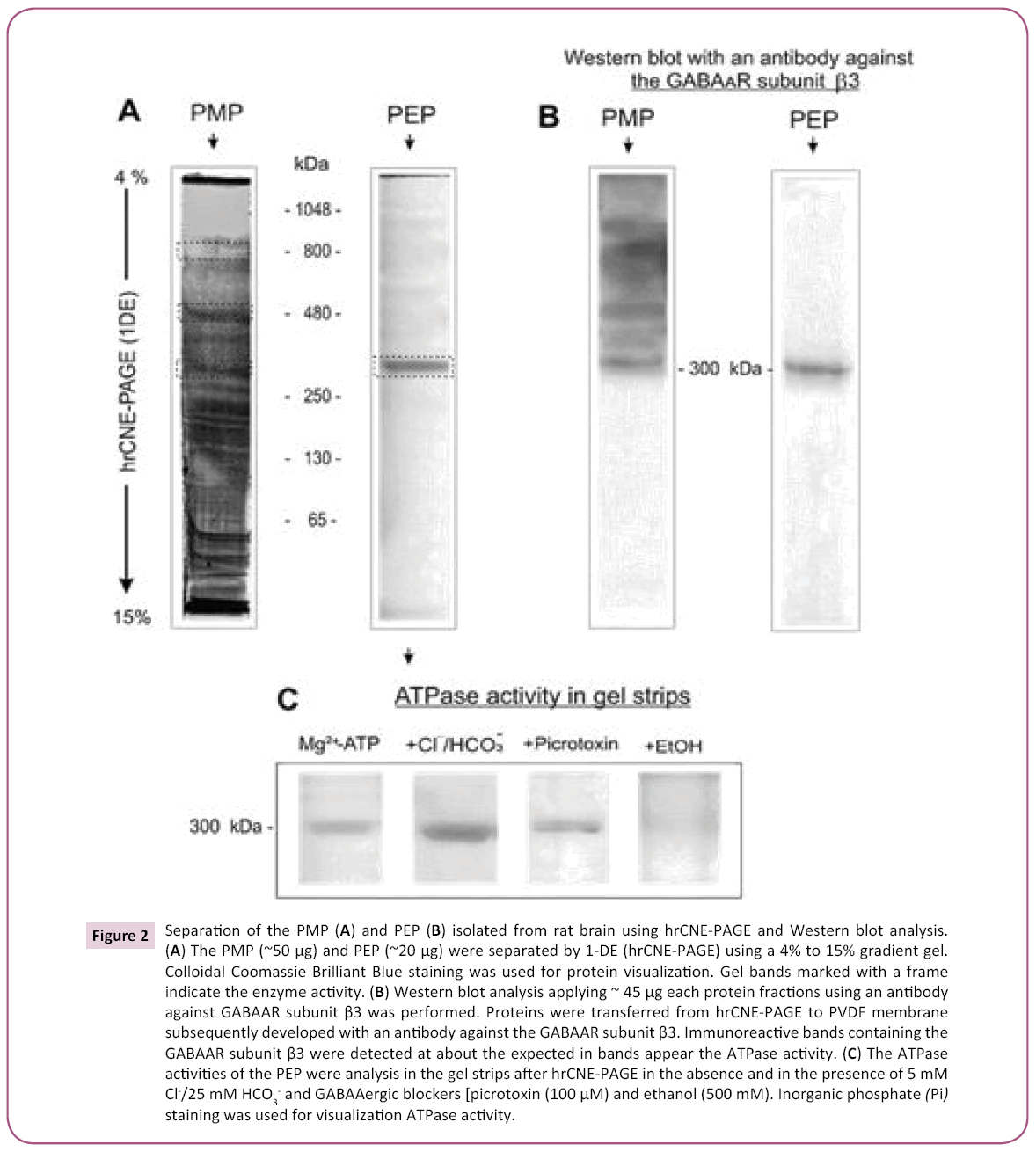
Figure 2: Separation of the PMP (A) and PEP (B) isolated from rat brain using hrCNE-PAGE and Western blot analysis. (A) The PMP (~50 μg) and PEP (~20 μg) were separated by 1-DE (hrCNE-PAGE) using a 4% to 15% gradient gel. Colloidal Coomassie Brilliant Blue staining was used for protein visualization. Gel bands marked with a frame indicate the enzyme activity. (B) Western blot analysis applying ~ 45 μg each protein fractions using an antibody against GABAAR subunit β3 was performed. Proteins were transferred from hrCNE-PAGE to PVDF membrane subsequently developed with an antibody against the GABAAR subunit β3. Immunoreactive bands containing the GABAAR subunit β3 were detected at about the expected in bands appear the ATPase activity. (C) The ATPase activities of the PEP were analysis in the gel strips after hrCNE-PAGE in the absence and in the presence of 5 mM Cl-/25 mM HCO3- and GABAAergic blockers [picrotoxin (100 μM) and ethanol (500 mM). Inorganic phosphate (Pi) staining was used for visualization ATPase activity.
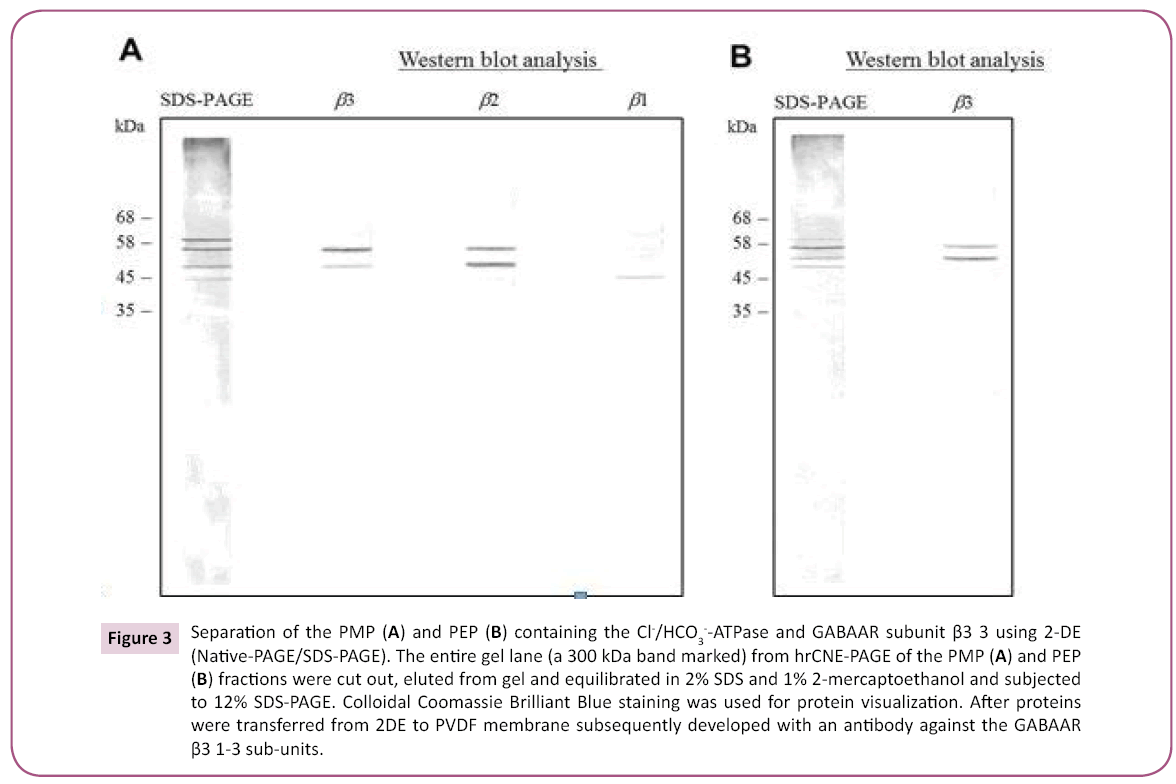
We further identified the nature of the protein complex comprising the ATPase activity by western blotting with an antibody against the GABAAR β3 subunit (Figure 2B). The PMP gel strips demonstrated an immunoreactivity to the GABAAR β3 antibody in bands with a molecular weight of 300-900 kDa, while the PEP demonstrated immunoprecipitation of the GABAAR β3 antibody with a single band at 300 kDa. The 2-D hrCNE-PAGE/ SDS-PAGE results were similar for both enzyme preparations, revealing four subunits with a molecular weight in the range of 45– 60 kDa (Figure 3A,B). The proteins resolved by SDS-PAGE were transferred to PVDF membranes for immunoblotting with antibodies against the GABAAR β1-3 subunits. Western blotting of 2-D electrophoresis gels for the PMP showed immunoreactivity with the GABAAR β2 and β3 subunits with similar molecular masses of 54 and 57 kDa, respectively (Figure 3A). The protein subunit that showed immunoreactivity with the GABAAR β1 subunit had an apparent molecular mass of 48 kDa. The PEP showed only two bands with molecular masses of 52 and 57 kDa that were immunoreactive with the GABAAR β3 antibody; no immunoreactivity was observed for the β1 and β2 antibodies (Figure 3B).
Figure 3: Separation of the PMP (A) and PEP (B) containing the Cl-/HCO3--ATPase and GABAAR subunit β3 3 using 2-DE (Native-PAGE/SDS-PAGE). The entire gel lane (a 300 kDa band marked) from hrCNE-PAGE of the PMP (A) and PEP (B) fractions were cut out, eluted from gel and equilibrated in 2% SDS and 1% 2-mercaptoethanol and subjected to 12% SDS-PAGE. Colloidal Coomassie Brilliant Blue staining was used for protein visualization. After proteins were transferred from 2DE to PVDF membrane subsequently developed with an antibody against the GABAAR β3 1-3 sub-units.
Phosphorylation and protein identification
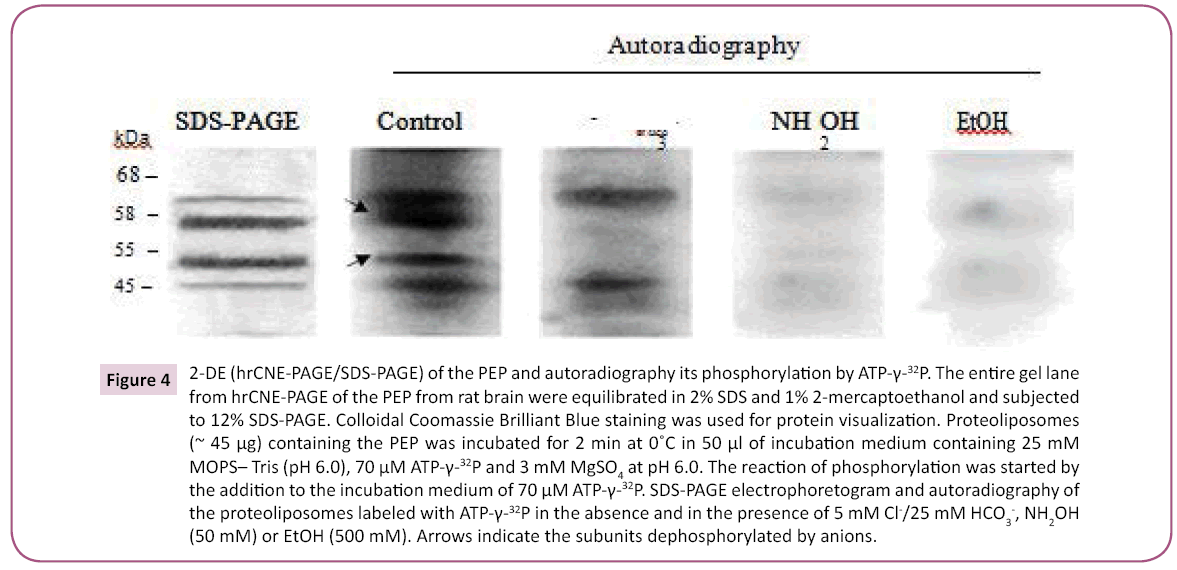
The available literature shows that direct phosphorylation of different transport P-type ATPases and Cl--ATPases can occur by [γ-32P]ATP in the presence of 3 mM Mg2+ [2]. In the present work, the enzyme preparations were reconstituted into proteoliposomes and tested for phosphorylation ability in the presence of [γ-32P] ATP and Mg2+ (3mM). Subsequent SDS-PAGE and autoradiography revealed the presence of phosphorylated proteins with molecular masses in the range of 45–60 kDa (Figure 4). The PEP showed protein bands with molecular weights of 52 kDa and 57 kDa that were dephosphorylated in the presence of 5 mM Cl-/25 mM HCO3-. The addition of EtOH (500 mM) and NH2OH (50 mM), a blocker of the high-energy phosphorylated (acyl phosphate) intermediate, resulted in complete inhibition of 32P incorporation into the studied peptide subunits.
Figure 4: 2-DE (hrCNE-PAGE/SDS-PAGE) of the PEP and autoradiography its phosphorylation by ATP-γ-32P. The entire gel lane from hrCNE-PAGE of the PEP from rat brain were equilibrated in 2% SDS and 1% 2-mercaptoethanol and subjected to 12% SDS-PAGE. Colloidal Coomassie Brilliant Blue staining was used for protein visualization. Proteoliposomes (~ 45 μg) containing the PEP was incubated for 2 min at 0˚C in 50 μl of incubation medium containing 25 mM MOPS– Tris (pH 6.0), 70 μM ATP-γ-32P and 3 mM MgSO4 at pH 6.0. The reaction of phosphorylation was started by the addition to the incubation medium of 70 μM ATP-γ-32P. SDS-PAGE electrophoretogram and autoradiography of the proteoliposomes labeled with ATP-γ-32P in the absence and in the presence of 5 mM Cl-/25 mM HCO3-, NH2OH (50 mM) or EtOH (500 mM). Arrows indicate the subunits dephosphorylated by anions.
Discussion
The GABAARs have diverse pharmacology due to the presence of the subunits that form the quaternary structure, as well as the allosteric binding sites for several different ligands (e.g., barbiturates, benzodiazepines, analgesics, anesthetics, and ethanol) [5,6,22,23]. Low concentrations of EtOH (<30 mM) activate and a high concentrations (>200 mM) inhibit the GABAAR function [24,25].
| List of symbols and abbreviations |
| GABAAR |
GABAA receptor |
| EtOH |
Ethanol |
| PMP |
Plasma membrane-enriched preparation |
| PEP |
Purified enzyme preparation |
| hrCNE-PAGE |
High resolution clear native Electrophoresis |
| SDS-PAGE |
Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
Table :List of symbols and abbreviations.
List of symbols and abbreviations.
Our previous cytochemical investigation determined that ethanol (>200 mM) exclusively inhibits the Mg2+-ATPase activity of the plasma membrane in elements of the GABAAergic dendrodendrite synapses, but has no effect on the enzyme activity in other subcellular organelles (primarily the mitochondria and nucleus) [26]. In the present study, the Cl-/HCO3--ATPase activity of the PEP was completely inhibited by EtOH (500 mM) and picrotoxin (100œ), indicating the availability binding sites for these ligands on the protein complex. Moreover, the stimulating of GABA effect on the Cl-/HCO3--ATPase activity was eliminated by these blockers, indicating a receptor-dependent pathway for mediator action on the protein complex.
Earlier literature described the use of proteomic techniques (native and denaturing gel systems) for the separation and identification of different enzymes, including ATPases [16,27,28]. Subsequent studies indicated that hrCNE, unlike BNE, was a more suitable technique for the separation and detection of membrane-bound enzyme complexes [16]. For example, the mitochondrial F1/F0-ATPase was identified by Pi staining of 1-D electrophoresis gels after incubation with Mg2+-ATP, in the presence and absence of the enzyme inhibitor oligomycin [16]. In our study, the sensitivity of the Cl-/HCO3--ATPase to ethanol inhibition in the gel strips after a hrCNE confirmed that the enzyme was a protein with a molecular weight of 300 kDa. These data are similar to previously reported results. A previous gelexclusion chromatography study also confirmed that the Cl-/ HCO3--ATPase complex isolated from rat brain membranes had a molecular weight of approximately 290–300 kDa [2].
The use of gel-based proteomic approaches (1/2/3-D electrophoresis) with western blotting is widely documented in the study of the GABAARs [16,17,29]. Thus, a 1-D BN-PAGE run of the PMP from the mouse hippocampus and subsequent western blot analysis revealed that proteins immunoreactive to the GABAAR β3 antibody migrated between 450 and 1236 kDa [17]. The authors of this previous study suggested that this range was due to glycosylation of the GABAAR β3 subunits, which offset in their electrophoretic mobility and resulted in apparent differences in molecular weight [17]. The PMP in the present study, after 1-D gel electrophoresis and western blotting, also showed immunoreactivity with the GABAAR β3 antibody in a wide range of molecular weights of 300–900 kDa. In addition, the PEP after 1-D electrophoresis showed that the spots corresponding to ATP hydrolysis and to specific immunoprecipitation with the GABAAR β3 antibody had similar molecular weights of 300 kDa. These results are similar to the properties described for the inhibitor receptors. Thus, the molecular mass of the native GABAAR complex from mammalian brain is approximately 300 kDa [5].
Previous studies have shown that GABAARs purified by affinity column or immunoaffinity columns containing antibodies against the γ2, β2, and β3 subunits electrophoretically manifested after SDS-PAGE in bands with molecular weights in the range of 48–60 kDa [29-31]. Subsequent western blots, as well as mass spectrometry analysis of the proteins in gel strips after 2-D BN/ SDS-PAGE and 3-D BN/SDS/SDS-PAGE, also showed the presence of the GABAAR β1-3 subunits [17,29-31]. In our work, 2-D CN/ SDS-PAGE and western blotting of the PMP showed the presence of the GABAAR β1-3 subunits, whereas the PEP demonstrated the presence only of the GABAAR β3 subunits, with molecular weights of 52 and 57 kDa. These data suggested that the ATPase was apparently associated with the GABAAR ensembles, which included β3 peptide subunits [6]. This assumption was also confirmed by the autoradiography results, which showed that all the peptide subunits of the PEP were phosphorylated by ATP and were dephosphorylated in the presence of hydroxylamine. However, only subunits with a molecular weight of 52 kDa and 57 kDa were dephosphorylated in the presence of anions and EtOH showed immunoreactivity to the GABAAR β3 antibody, indicating that they belonged to the same protein complex.
The human and mammalian brains contain at least fifteen GABAAR assemblies that differ in subunit composition, regional and cellular distribution, and pharmacology [6,22,23]. However, most of the GABAAR ensembles (~ 60%) that are present in the brain have the α1/2, β2/3, and γ2 subunits [6]. The most common GABAAR compositions containing the β3 subunit are α3β3γ2 and sα2β3γ2. The literature also shows that some GABAAR ensembles containing the β3 subunit are selectively sensitive to ethanol [2]. For example, the mammalian brain contains an ethanol-sensitive GABAAR assembly composed of α4β3δ or α6β3δ subunits [33]. One possibility is that this enzyme is associated with this type of inhibitory receptor.
Conclusion
The proteomic approach and western blotting analysis described here allowed the demonstration of the presence of GABAAR β3 subunits within a purified preparation of the Cl-/HCO3-- ATPase. The molecular weight and subunit composition of the investigated ATPase were analogous to the molecular properties of the GABAAR. The phosphorylation results also support this finding, confirming not only the functional but also the structural coupling of the enzyme with the inhibitory receptors. Further application of this gel-based proteomic technique, together with mass spectrometry and analysis of site-specific phosphorylation, should reveal the nature of the GABAAR ensembles associated with this ATPase and should allow identification of the catalytic subunit. The use of a proteomic approach can therefore be viewed as a major step for clarifying the structural association of Cl-/HCO3--ATPase with GABAAR ensembles and in the search for analytical methods to work on the enzyme in mammalian brain in health and disease.
References
- Menzikov SA, Karpova MN, Kalinina MV (2011) Effect of HCO3- ions on the ATP-dependent GABAA receptor-coupled Cl- channel in rat brain plasma membranes. Bull ExpBiol Med 152: 38–42.
- Menzikov SA (2013) Neuronal multifunctional ATPase. Biophys Rev Lett8: 213–227.
- Menzikov SA (2017) Isolation, purification, and partial characterization of a membrane-bound Cl/HCO3-activated ATPase complex from rat brain with sensitivity to GABAAergic ligands. Prep BiochemBiotechnol 47: 151-157.
- Staley KJ, Proctor WR (1999) Modulation of mammalian dendritic GABAA receptor function by the kinetics of Cl- and HCO3-transpþrt. J PhysiolLond 519: 693–712.
- Michels G, Moss SJ (2007) GABAA receptors: Properties and trafficking. Crit Rev BiochemMolBiol 42: 3-14.
- Nutt D (2006) GABAA Receptors: Subtypes, regional distribution, and function. J Clin Sleep Med. 2: 7-11.
- Sigel E, Steinmann ME (2012) Structure, function, and modulation of GABA(A) receptors.J BiolChem 287: 40224-40231.
- Jensen ML, Timmermann DB, Johansen TH, Schousboe A, Varming T, et al. (2002) The beta subunit determines the ion selectivity of the GABAA receptor. J BiolChem 277: 41438-41447.
- Connolly CN, Wooltorton JR, Smart TG, Moss SJ (1996) Sub-cellular localization of gamma-aminobutyric acid type A receptors is determined by receptor beta subunits. ProcNatlAcadSci USA 93: 9899-9904.
- Davis MJ, Wu X, Nurkiewicz TR, Kawasaki J, Gui P, et al. (2001) Regulation of ion channels by protein tyrosine phosphorylation. Am J Physiol Heart CircPhysiol 281: 1835– 1862.
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, et al. (2000) Active transport by ATP-powered pumps. Molecular Cell Biology. (4th edn), New York. W. H. Freeman
- Vithlani M, Terunuma M, Moss SJ (2011) The dynamic modulation of GABA(A) receptor trafficking and its role in regulating the plasticity of inhibitory synapses. Physiol Rev 91: 1009-1022.
- Stelzer A, Kay AR, Wong RK (1988) GABAA-receptor function in hippocampal cells is maintained by phosphorylation factors. Science 241: 339-341.
- Watson GB, Salgado VL (2001) Maintenance of GABA receptor function of small-diameter cockroach neurons by adenine nucleotides. Insect BiochemMolec Biology 31: 207–212.
- Cupello A (2003) Neuronal transmembrane chloride electrochemical gradient: A key player in GABAA receptor activation physiological effect. Amino Acids 24: 335–346.
- Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6: 1215-1225.
- Kang SU, Heo S, Lubec G (2011) Mass spectrometric analysis of GABAA receptor subtypes and phosphorylations from mouse hippocampus. Proteomics 11: 2171-2181.
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principal of protein-dye binding. Anal Biochem 72: 248–254.
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage. Nature 227: 680–685.
- Gonchar MV, Monastyrskiy VA (1982) Micromethod determination of inorganic phosphate in the blood. Laboratory work 1: 29-30.
- Menzikov S, Karpova M,Kuznetsova L,Klishina, N (2015) GABAA-coupled Cl-/HCO3-- ATPase from plasma membrane of the rat brain: role of HCO3- in the enzyme activation, Advances in Enzyme Research 3: 9-18.
- Sigel E, Steinmann ME (2012) Structure, function, and modulation of GABA(A) receptors. J BiolChem 287: 40224-40231.
- Sieghart W (2015) Allosteric modulation of GABAA receptors via multiple drug-binding sites. AdvPharmacol 72: 53-96.
- Wakita M, Shin MC, Iwata S, Nonaka K, Akaike N (2012) Effects of ethanol on GABA(A) receptors in GABAergic and glutamatergic presynaptic nerve terminals. J PharmacolExpTher 341: 809-819.
- Xie Z, Lia G, Yea JH (2013) Acute effects of ethanol on GABAA and glycine currents in the lateral habenula neurons of young rats. Open Journal of Neuroscience. 3-5.
- Menzikov SA, Ruzhinskaia NN, Menzikova OV (2000) Mg2+-ATPase in the fish brain and its ultrastructural localization. ZhEvolBiokhimFiziol 36: 263-267.
- Rabilloud T, Lelong C (2011) Two-dimensional gel electrophoresis in proteomics: a tutorial.JProteomics 74: 1829-1841.
- Krause-Buchholz U,Becker JS,Zoriy M,Pickhardt C,Przybylski M, et al. (2006) Detection of phosphorylated subunits by combined LA–ICP–MS and MALDI–FTICR–MS analysis in yeast mitochondrial membrane complexes separated by blue native/SDS-PAGE.i J Mas Spectr 248: 56–60.
- Chen ZW, Fuchs K,Sieghart W, Townsend RR, Evers AS (2012) Deep amino acid sequencing of native brain GABA(A) receptors using high-resolution mass spectrometry.Mol Cell Proteomics 11: 111-445.
- Gavish M, Snyder SH (1981) Gamma-aminobutyric acid and benzodiazepine receptors: Co-purification and characterization. ProcNatlAcadSci USA. 78: 1939–1942.
- Jechlinger M, Pelz R, Tretter V, Klausberger T, Sieghart W (1998) Subunit composition and quantitative importance of hetero-oligomeric receptors: GABAA receptors containing alpha6 subunits. J Neurosci 18: 2449-2457.
- Van Skike CE, Diaz-Ganados JL, Matthews DB (2015) Chronic intermittent ethanol exposure produces persistent anxiety in adolescent and adult rats. Alcohol ClinExp Res 39: 262-271.
- Wallner M, Hanchar HJ, Olsen RW (2014) Alcohol selectivity of β3-containing GABAA receptors: Evidence for a unique extracellular alcohol/imidazobenzodiazepine Ro15-4513 binding site at the α+β- subunit interface in αβ3δ GABAA receptors. Neurochem Res 39: 1118-1126.