Rocío Espada1*, Diego U Ferreiro1 and Gonzalo Parra R2
1Protein Physiology Laboratory, Departamento de Química Biológica, Iquibicen-Conicet, Universidad de Buenos Aires, Argentina
2Quantitative and Computational Biology Group, Max Planck Institute for Biophysical Chemistry, Goettingen, Germany
Corresponding Author:
Rocío Espada
Protein Physiology Laboratory
Departamento De Química Biológica
Iquibicen-Conicet, Universidad De
Buenos Aires, Argentina
Tel: +49 551 201-2891
E-mail: parra.gonzalo@gmail.com
Received date: February 21, 2017; Accepted date: March 24, 2017; Published date: March 27, 2017
Citation: Espada R, Ferreiro DU, Parra RG. The Design of Repeat Proteins: Stability Conflicts with Functionality. Biochem Mol Biol J. 2017, 3:1. doi: 10.21767/2471-8084.100031
Keywords
Repeat proteins; Protein folding; Designed proteins; Protein stability; Local frustration
Introduction
The more sequences, structures and functions scientists have discovered about proteins, the more they have dreamed about being able to design and engineer new ones for specific aims. Globular proteins have intricate topologies where residues that are far in sequence come close in space upon folding. This complicates the analysis of how sequence perturbations are propagated to the structure, the overall dynamics and finally their impact on protein function. Repeatproteins could simplify this problem. Repeat proteins are constituted by a variable number of fundamental structural elements that are repeated in tandem along a longitudinal axis. They can adopt different shapes according to the geometrical and symmetry relation in between the repeating modules [1,2]. Residue-residue interactions on these molecules are mostly confined within each repetition or to the interfaces between adjacent ones and in principle local perturbations are not propagated to distant regions in the structure. In their natural context, repeat proteins are frequently found mediating protein-protein interactions, with a specificity rivaling that of antibodies [3-5]. For these reasons, it is not unexpected that several groups have used them as scaffolds for the design of protein interactors with notable success [6].
Designed repeat proteins have high thermal stabilities compared to the natural occurring ones as reviewed in [7]. While high stability might be a desired feature for obtaining well expressed and foldable polypeptides, most natural proteins are marginally stable [8]. It has been suggested that marginal stability could favor protein functionality since it would be correlated with increased flexibility [9], and it has been suggested that this could increase "functionality" [10,11]. Proteins' exploration of conformational states is now recognized as fundamental to protein function [12,13] to transition between the different conformers that constitute their native state and hence be able to accomplish their function [14].
If proteins are naturally marginally stable, is the maximization of stability the right coordinate for protein design? The energy landscapes theory for protein folding [15] explains that proteins minimize their internal conflicts as they fold towards structures that are more similar to their native state. This global minimization of the internal energy is known as "the principle of minimal frustration" and it is a consequence of the cooperativity that exists among native interactions. From all possible polypeptides, natural sequences constitute the set for which most native interactions are more favourable than any other possible interaction within the polypeptide chain [15]. Although minimized, the principle of minimal frustration does not imply that some energetic conflicts cannot remain in the native states. Even more, it has been shown that, in average, 10% of the total interactions in a monomeric, foldable protein structure are in conflict with their local environment [16]. These "highly frustrated" interactions, that have been kept along the evolutionary history of proteins [17] are crucial for several functional aspects of natural proteins [18].
Current methods to design repeat proteins rely on the implicit or explicit maximization of stability, while nature supposedly cares about proteins being functional in their environment. We discuss here the importance of energetic conflicts in natural occurring repeat proteins and why they should be taken into account for improving their use as scaffolds for protein design.
Repeat Proteins Design: Regular and Highly Stable Molecules
One of the most successful strategies for designing proteins is the so called "consensus-based" approach which implicitly exploits the evolutionary information from a set of evolutionary related proteins. Starting from a multiple sequence alignment of homologous protein domains, the most probable amino acid at each position is selected to create a consensus sequence which can be synthesized [7]. In the case of repeat proteins, multiple sequence alignments of individual repeats are used for constructing the consensus sequence. Multiple copies of this consensus repeat can be concatenated in order to generate consensus repeat proteins of different lengths. Several groups have successfully used Ankyrin repeat proteins (ANK) for consensus-design [19-22]. Peng and coworkers [19] generated proteins composed of one to four identical Ankyrin repeat units (1ANK, 2ANK, 3ANK, 4ANK) and showed that ANK3 and ANK4 are foldable with high thermal stabilities. These results demonstrated that repeat consensus sequences contain all the information, within repeats and between adjacent repeats interactions, needed to fold a molecule with the Ankyrin repeat architecture. Pluckthun and coworkers on their side [20] generated another consensus design, leaving 6 positions free to be randomized in order to generate libraries in which specific binders could be found. These DARPins (Designed Ankyrin Repeat Proteins) have been very successful. The structure of a DARPin with 5 repeats, E3_5, was solved showing to be very regular and well packed [23]. Regardless on the differences on their design strategies 3ANK, 4ANK and E3_5 are able to correctly fold with higher stabilities than any other natural ANK. Other repeat protein families were used as scaffolds for consensus-based design like Leucine Rich Repeats (LRR), Tetratricopeptide Repeats (TPR), Armadillo (ARM) and HEAT Repeats [24]. In recent years, variations of consensus designed repeat proteins permitted to recognize a diversity of specific targets [6]. Nevertheless, the rigidity of these proteins at their binding surface limits the diversity of molecules that can be targeted by this extremely stable scaffold, with some strategies being applied to compensate this as for example the inclusion of insertions of variable lengths in some of the repeats [25].
In a recent work, we studied the energetics of the entire ANK family [26]. By using an algorithm called Frustratometer [27,28] we analyzed the frustration patterns in all ANKs with known structure. The Frustratometer compares the native interaction energy of a given interaction with the one that would be found by placing different residues in the same native location or by creating a different environment for the interacting pair. When an interaction is more favourable than most of the alternatives it is considered to be minimally frustrated. If, on the contrary, most of the alternatives are more favourable than the native energy, the interaction is considered to be highly frustrated. Interactions whose energy values cannot be distinguished from the alternatives mean energy values are considered to be neutral. We observed that energetic conflicts (highly frustrated interactions) were not randomly distributed but specifically located at binding sites, insertions and regions close to deletion points [26]. When comparing the sequence and the structural energy conservation at each position, we found that the more similar to the consensus a residue is, the more it contributes to the repeat stability by establishing energetically favourable, i.e., minimally frustrated, interactions both within the repeat and with the neighboring ones. Interestingly, positions that can be randomized in the DARPins design are not included in the set of residues that are important for the repeat stability. Residues that are marked as "functional" tend to be responsible for disrupting the periodicity in the repeat array and are enriched in energetic conflicts, while those that are maintained to be similar to the canonical ANK structure tend to favour protein stability.
In Figure 1, we show the frustration patterns for different protein structures corresponding to designed repeat proteins (first and second rows) and for natural repeat proteins (third row). When comparing the frustration patterns for the full consensus Ankyrin repeat protein 4ANK (Figure 1A) and the natural one IκBα (Figure 1H), which is an inhibitor of NF-κB, it appears that the first one has a lower proportion of conflictive interactions (red lines). IκBα is known to be partially folded and only consolidates its structure upon interacting with NF-κB [29,30]. On the other hand, 4ANK is a full consensus Ankyrin repeat protein, that shows a neat minimally frustrated pattern, consistent with the overall observation that residues that maximize the similarity to the consensus sequence are responsible for maximizing repeat stability [26].
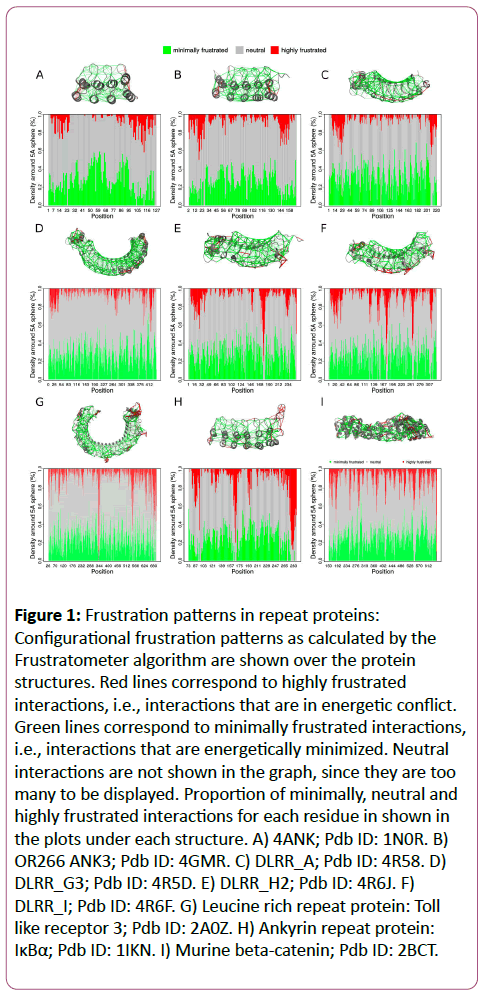
Figure 1: Frustration patterns in repeat proteins: Configurational frustration patterns as calculated by the Frustratometer algorithm are shown over the protein structures. Red lines correspond to highly frustrated interactions, i.e., interactions that are in energetic conflict. Green lines correspond to minimally frustrated interactions, i.e., interactions that are energetically minimized. Neutral interactions are not shown in the graph, since they are too many to be displayed. Proportion of minimally, neutral and highly frustrated interactions for each residue in shown in the plots under each structure. A) 4ANK; Pdb ID: 1N0R. B) OR266 ANK3; Pdb ID: 4GMR. C) DLRR_A; Pdb ID: 4R58. D)DLRR_G3; Pdb ID: 4R5D. E) DLRR_H2; Pdb ID: 4R6J. F)DLRR_I; Pdb ID: 4R6F. G) Leucine rich repeat protein: Tolllike receptor 3; Pdb ID: 2A0Z. H) Ankyrin repeat protein:IκBα; Pdb ID: 1IKN. I) Murine beta-catenin; Pdb ID: 2BCT.
Recently, a new methodology for designing repeat proteins has been developed by Baker and co-workers [31] that integrates the Rosetta de novo structure generation and design methodology combined with protein family-based sequence and structural information. Starting from poly-valine protein backbones, several models, composed of idealized repetitive modules are obtained for which a Rosetta energy function, supplemented with family-specific structural constraints, is optimized. This strategy has been successful for the design of novel repeat-protein topologies [31]. In Figure 1B, we show the structure for ANK3 (OR266), designed by the mentioned method, which consists of 3 internal idealized repeats and adapted versions for the terminal ones (Caps). We observe that ANK3 has a larger proportion of highly frustrated interactions, compared to the full consensus ANK4 in (Figure 1A) not only in the Caps, but also in the internal repeats. This strategy was extended later on [32], using the Leucine Rich Repeat (LRR) family as a scaffold, for designing repeat proteins for which the curvature of the array could be rationally modified. The curvature is essential to tune the complementarity between the repeat protein and its target. The methodology consists of 3 steps: 1) Design of a set of idealized self-compatible building block modules, 2) design of junction modules that connect adjacent building blocks from 1, and 3) combination of building and junction blocks in order to generate a protein with a specific desired overall curvature. Several LRR constructs were obtained following this approach by combination of idealized LRR modules of different lengths (LRR_L, where L is the length of the repeat module) building modules and junction modules. In Figure 1C we show the frustration pattern for DLRR_A which is constituted by LRR_22 idealized modules which has frustrated interactions on the terminal regions, and a lower proportion on the internal repeats. In Figure 1D we show the frustration patterns for DLRR_G3, which is constituted by fusion of LRR_24 and LRR_28 idealized modules. It can be seen that there is a central region around residue 280, corresponding to the interfaces between the LRR_24 and the LRR_28 modules, that has a larger proportion of highly frustrated interactions compared to the rest of internal repeats.
Natural LRRs, usually contain non repetitive "irregular" regions. For example, the Toll-like receptor 3, has a repeat, located between residues 532 and 563, with an inserted loop extending the repeat length to 32 residues (LRR_32) respect the canonical LRR_24 idealized repeats (Fig. 1G). The frustration patterns for DLRR_H2 containing one LRR_32 module in combination with LRR_24 modules and DLRR_I that contains two consecutive LRR_32 modules are shown in (Figures 1E and 1F), respectively. We observe that in both the cases, the designed proteins and the Toll-like receptor 3, the regions where the wedges are located have a higher proportion of highly frustrated interactions. We have shown for the ANK protein family, that insertions within and in between adjacent repeats are enriched in this type of energetic conflicts [26], most likely because of functional constraints like surface adaptation to bind the target, as suggested in the case of LRRs or for dynamic and regulatory reasons like in the IκBα case [29,33].
Discussion
Repeat proteins are excellent models for studying the sequence-structure-dynamics-function relationships because of their simplified topologies that minimize long-range interactions within the polypeptide chains. For this same reason, several groups have used them as scaffolds for protein design. Designed repeat proteins have higher thermal stabilities than their natural counterparts. This higher stability is a consequence of the implicit maximization of the gap between the folded and unfolded states [26] in the case of consensus-based design approaches [19,20] and the minimization of an energetic function in the case of the de novo design method based on Rosetta [31] which is another strategy to maximize the aforementioned gap [34]. Although quite successful, maximizing stability in designed proteins lead to constructs with several limitations as a consequence of their lack of flexibility. Energetic analysis on the ANK family showed that natural ANKs contain much more energetic conflicts than designed ones, mainly located in non-repetitive irregularities, i.e., insertions and deletions, as well as in those residues involved in protein-protein interactions [26]. These nonperiodic elements, that tend to disrupt the propagation of symmetry on the global structure [35], are also responsible for energetic conflicts when added to synthetic constructs like in the case of DLRR_H2 and DLRR_I (Figure 1E and 1F). The high thermal stability of highly periodic constructs like 4ANK, ANK3 or DLRR_A is affected by the presence of symmetry disrupting elements, as in DLRR_H2 and DLRR_I. Natural repeat proteins, exquisitely combine their modular structure with several sources of instability to modulate the internal stability of repeats as well as the repeat-repeat interaction energetics which may be related to their biological functions [36,37]. This plasticity allows them to adapt their interfaces to optimize their binding properties to a given target with high affinities and specificities. This was explicitly shown when analyzing all ANKs structures that have been co-crystallized as being part of a quaternary complex, where much of the energetic conflicts, present at the binding interfaces, are released once the complex is formed [26]. It is not surprising then that designed repeat proteins that contain considerably less energetic conflicts (Figure 1A-1F) are not as good at recognizing their targets as the natural ones (Figure 1G-1I) which in turn can display complex dynamic behaviours. By getting rid of most of their energetic conflicts, while maximizing their stability, designed repeat proteins may not keep enough local instabilities to become more stable when complexing with their interactors as natural ones and hence promote and drive the recognition process.
Conclusion
Protein design strategies using repeat proteins as scaffolds have made impressive advances with many already on going applications [6] but still, the main paradigm consists on the obtention of highly stable molecules which lead to some limitations when more versatile binders are desired. These designed proteins are well behaved to favour expression and folding that can be further optimized for binding a specific target [31,38]. On their side, most natural proteins are marginally stable and biology has taken advantage of this property and may have used it as a "spandrel" to develop protein function [39]. The trade-off in between stability and protein function has long been discussed in [40] and references therein. In many cases, like in enzymes, the change of key functional residues, like the catalytic ones, which often possess unfavorable energetics, can dramatically increase stability at the expense of activity [41,42]. In some repeat proteins stabilization of the repeat array by consensus residues have indirect consequences. It was shown that stabilization of the 6th repeat on IkBa protein promotes the ordering of the PEST region, increasing its resistance to degradation [43] and functional properties [44] and has been related to be relevant for several types of cancer, autoimmune diseases and other pathologies [45]. The challenge in the design of repeat proteins is to understand how to modulate the stabilityfunction trade-off so new functions can be imprinted on them in a rational manner.
The increasing availability of sequences derived from metagenomic studies along with the development and adaptation of specific computational tools [35,46-48] and databases [2,49] assists in the study of repeat proteins properties and classification [50]. This in turn will facilitate to tackle the different challenges repeat proteins pose [51] and help to dissect the details in each of their sequence-structuredynamics- function relationships. We believe that a deeper understanding of natural repeat protein families and how they conjugate stability and energetic conflicts will provide new insights for improving the design of these molecules and the obtention of better protein-protein interactors.
References
- Kajava AV(2012) Tandem repeats in proteins: From sequence to structure. Journal of Structural Biology, 179: 279–288.
- Domenico TD, Potenza E,Walsh I, Gonzalo PR,Giollo M, et al. (2013) RepeatsDB: A database of tandem repeat protein structures. Nucleic Acids Research p: gkt-1175.
- Sedgwick SG,Smerdon SJ (1999) The ankyrin repeat: A diversity of interactions on a common structural framework. Trends in Biochemical Sciences 24: 311–316.
- Kobe B,Kajava AV (2001) The leucine-rich repeat as a protein recognition motif. Current Opinion in Structural Biology11: 725–732.
- Blatch GL,Lässle M (1999) The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays21: 932–939.
- Plückthun A(2015) Designed ankyrin repeat proteins (darpins): Binding proteins for research, diagnostics, and therapy. Annual Review of Pharmacology and Toxicology, 55: 489–511.
- Porebski BT, Buckle AM(2016) Consensus protein design. Protein Engineering Design and Selection 29: 245–251.
- Privalov PL,Khechinashvili NN (1974) A thermodynamic approach to the problem of stabilization of globular protein structure: A calorimetric study. Journal of molecular biology, 86: 665–684.
- Zavodszky P, Kardos J, Svingor Á, Petsko GA (1998) Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proceedings of the National Academy of Sciences, 95: 7406–7411.
- Wright PE,Dyson HJ (1999) Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. Journal of Molecular Biology 293: 321–331.
- Dyson HJ, Wright PE(2002) Coupling of folding and binding for unstructured proteins. Current Opinion in Structural Biology12: 54–60.
- Hegler JA, Weinkam P, Wolynes PG(2008) The spectrum of biomolecular states and motions. HFSP Journal, 2: 307–313.
- Boehr DD, Nussinov R, Wright PE(2009) The role of dynamic conformational ensembles in biomolecular recognition. Nature chemical biology, 5: 789–796.
- Hemanth GRVV, Gosavi S(2016) Using the folding landscapes of proteins to understand protein function. Current Opinion in Structural Biology. 36: 67–74.
- Bryngelson JD,Wolynes PG (1987) Spin glasses and the statistical mechanics of protein folding. Proceedings of the National Academy of Sciences. 84: 7524–7528.
- Ferreiro DU, Hegler JA, Komives EA, Wolynes PG(2007) Localizing frustration in native proteins and protein assemblies. Proceedings of the National Academy of Sciences. 104: 19819–19824.
- Wolynes PG(2015) Evolution, energy landscapes and the paradoxes of protein folding. Biochimie 119: 218–230.
- Ferreiro DU, Komives EA, Wolynes PG(2014) Frustration in biomolecules. Quarterly Reviews of Biophysics 47: 285–363.
- Mosavi LK, Minor DL, Zheng-Yu P (2002) Consensus-derived structural determinants of the ankyrin repeat motif. Proceedings of the National Academy of Sciences, 99: 16029–16034.
- Binz HK, Stumpp MT, Forrer P, Amstutz P, Plückthun A(2003) Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. Journal of Molecular Biology 332: 489–503.
- Tripp KW, Barrick D(2003) Folding by consensus. Structure 11: 486–487.
- Main ERG, Jackson SE, Regan L(2003) The folding and design of repeat proteins: Reaching a consensus. Current Opinion in Structural Biology 13: 482–489.
- Kohl A, Binz HK, Forrer P, Stumpp MT, Plückthun A, et al. (2003) Designed to be stable: Crystal structure of a consensus ankyrin repeat protein. Proceedings of the National Academy of Sciences 100(4): 1700–1705.
- Boersma YL,Plückthun A(2003) Darpins and other repeat protein scaffolds: Advances in engineering and applications. Current Opinion in Biotechnology 22: 849–857.
- Schilling J, Schöppe J, Plückthun A(2014) From darpins to loop darpins: novel loopdarpin design allows the selection of low picomolar binders in a single round of ribosome display. Journal of Molecular Biology426: 691–721.
- Parra RG, Espada R, Verstraete N, Ferreiro DU (2015) Structural and energetic characterization of the ankyrin repeat protein family. PLoSComputBiol 11: e1004659.
- Jenik M, Parra RG, Radusky LG, Turjanski A, Wolynes PG, et al. (2012) Protein frustratometer: A tool to localize energetic frustration in protein molecules. Nucleic Acids Research p:gks-447.
- Parra RG, Schafer NP, Radusky LG, Tsai MY, GuzovskyAB, et al. (2016) Protein frustratometer 2: A tool to localize energetic frustration in protein molecules, now with electrostatics. Nucleic Acids Researchp: gkw-304.
- Truhlar SME, Mathes E, Cervantes CF,Ghosh G(2008) Pre-folding IκBα alters control of NF-κBsignaling. Journal of Molecular Biology, 380: 67–82.
- Dyson HJ,Komives EA(2012) Role of disorder in IκB–NF-κB interaction. IUBMB Life64: 499–505.
- Parmeggiani F, Po-Ssu H, Vorobiev S, Xiao R, Park K, et al. (2015) A general computational approach for repeat protein design. Journal of Molecular Biology 427: 563–575.
- Park K, Shen BW, Parmeggiani F, Po-Ssu H, Stoddard BL, et al. (2015) Control of repeat-protein curvature by computational protein design. Nature Structural & Molecular Biology 22: 167–174.
- Dembinski H, Wismer K, Balasubramaniam D, Gonzalez HA, Alverdi V, et al. (2014) Predicted disorder-to-order transition mutations in IκBαdisrupt function. Physical Chemistry &Chemical Physics, 16: 6480–6485.
- Fleishman SJ, Baker D(2012) Role of the biomolecular energy gap in protein design, structure, and evolution. Cell 149: 262–273.
- Parra RG, Espada R, Sánchez IE, Sippl MJ, Ferreiro DU (2013) Detecting repetitions and periodicities in proteins by tiling the structural space. The Journal of Physical Chemistry B, 117: 12887–12897.
- Ferreiro DU, Komives EA(2007) The plastic landscape of repeat proteins. Proceedings of the National Academy of Sciences 104: 7735–7736.
- Ferreiro DU, Walczak AM, Komives EA, Wolynes PG (2008) The energy landscapes of repeat-containing proteins: topology, cooperativity, and the folding funnels of one-dimensional architectures. PLoSComputBiol 4: e1000070.
- Jost C,Plückthun A (2014) Engineered proteins with desired specificity: Darpins, other alternative scaffolds and bispecificIGgs. Current Opinion in Structural Biology27: 102–112.
- Taverna DM, Goldstein RA(2002) Why are proteins marginally stable? Proteins: Structure, Function, and Bioinformatics 46: 105–109.
- Tokuriki N, Stricher F, Serrano L, Tawfik DS (2008) How protein stability and new functions trade off. PLoSComputBiol 4: e1000002.
- Beadle BM,Shoichet BK (2002) Structural bases of stability–function tradeoffs in enzymes. Journal of Molecular Biology321: 285–296.
- Meiering EM, Serrano L, Fersht AR(1992) Effect of active site residues in barnase on activity and stability. Journal of Molecular Biology, 225: 585–589.
- Cervantes CF, Handley LD, Shih-Che S, Dyson HJ, Komives EA (2013) Long-range effects and functional consequences of stabilizing mutations in the ankyrin repeat domain of IκBα. Journal of Molecular Biology 425: 902–913.
- Dembinski HE, Wismer K, Vargas JD, Suryawanshi GW, Kern N,et al. (2017) Functional importance of stripping in NFΚbsignaling revealed by a stripping-impaired IκBα mutant. Proceedings of the National Academy of Sciences p: 201610192.
- Courtois G, Gilmore TD(2006) Mutations in the NF-κBsignaling pathway: implications for human disease. Oncogene 25: 6831–6843 2006.
- Espada R,Parra RG, Mora T, Walczak AM, Ferreiro DU (2015)Capturing coevolutionary signals in repeat proteins. BMC Bioinformatics, 16: 207.
- Hirsh L, Piovesan D, Paladin L,Tosatto SCE(2016) Identification of repetitive units in protein structures with repeated. Amino acids, 48: 1391–1400.
- Turjanski P, Parra RG, Espada R, Becher V, Ferreiro DU (2016) Protein repeats from first principles. Scientific Reports, 6.
- Paladin L, Hirsh L, Piovesan D, Andrade-Navarro MA(2016)RepeatsDB 2.0: Improved annotation, classification, search and visualization of repeat protein structures. Nucleic Acids Research p:gkw-1136.
- Paladin L,Tosatto SCE(2015) Comparison of protein repeat classifications based on structure and sequence families. Biochemical Society Transactions 43: 832–837.
- Espada R, Parra GR, Sippl MJ, Mora T, Walczak AM, et al. (2015) Repeat proteins challenge the concept of structural domains. Biochemical Society Transactions43: 844–849.


