Research Article - (2018) Volume 8, Issue 6
Somayeh Khodayari1* and Farshid Kafilzadeh2
1Young Researchers and Elite Club, Jahrom Branch, Islamic Azad University, Jahrom, Iran
2Department of Microbiology, Jahrom Azad University, Jahrom, Iran
Corresponding Author:
Khodayari S
Young Researchers and Elite Club
Jahrom Branch, Islamic Azad University, Jahrom, Iran
Tel: 0098 9177396265
E-mail: Khodayari.skho@gmail.com
Received Date: Oct 24, 2018; Accepted Date: Nov 16, 2018; Published Date: Nov 26, 2018
Citation: Khodayari S, Kafilzadeh F (2018) Separating Keratinase Producer Bacteria from the Soil of Poultry Farms and Optimization of the Conditions for Maximum Enzyme Production. Eur Exp Biol Vol. 8 No. 6:35. doi:10.21767/2248-9215.100076
Copyright: © 2018 Khodayari S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Feather is considered one of the environmental pollutant factors that can be hydrolyzed by bacteria and fungi. About 90% of the full weight of a feather consists of keratin. The structure of feather is very difficult to break down. Some bacteria in the presence of keratin-contained substrates are able to produce the keratinase enzyme to hydrolyze keratin.
A total of 15 soil samples were collected from the poultry farms around Marvdasht city. 7 bacterial strains were grown in feather filled environment. 5 isolates that showed a clear analysis were selected and identified using biochemical tests and molecular methods. The bacterial DNA contents were sequenced and were assigned certain numbers in GenBank as new strains. Then, every fifth bacteria were also evaluated for the production of keratinase.
All strains belonged to different strains of Bacillus. Five strains were able to completely degrade feather. Bacillus cereus SKH1 had the maximum enzyme activity, 17.12 (Unit/ml/min).
Different strains of Bacillus in this study showed the ability to produce keratinase in the presence of keratin. Keratinase measurement showed that all 5 strains are potentially able to treat feather-contained waste.
Keywords
Keratinase enzyme; keratin hydrolyzer bacteria; Bacillus cereus SKH1
Introduction
Keratin is an insoluble macromolecule with high stability and low speed of decomposition and is found in hair, feather, nail, fur, beak, and horn [1,2]. Keratins are divided into two main groups: α-keratin and β-keratin [3]. In both groups, Keratin fibrils are wrapped together in parallel to form micro and macro dimer structures with high mechanical stabilities [4,5]. Feather is made from pure Keratin and is very insoluble and difficult to decompose [6]. Annually, billions of chickens are consumed around the world and about 8.5 billion tons of chicken feathers are produced [7]. Feather consists about 5% to 7% of the chicken weight and keratin consists about 90 percent of the feather weight [8,9]. Feathers are used to fill the earth, burning or burying and these processes, produce problems with the storage, transportation, and disposal of ash and greenhouse gas emissions [6,10]. In addition, feather, as a byproduct, causes various diseases in humans, including chlorosis and mycoplasmosis [10]. The high content of protein in the keratincontained waste can become a good source of protein and amino acids using recycling mechanisms [2]. Feather recycling can provide low-cost protein-rich foods for other animals. Although poor digestion of keratin is a big problem in the recycling of feather, using hydrothermal and chemical treatment with high temperature and high pressure, may help metabolize keratin easily and this coincides with the loss of some essential amino acids such as methionine, lysine, and tryptophan, which lowers the nutritional value of keratin. Thus, thermal and chemical treatments do not retain enough nutritional content [11,12]. Here, microorganisms with keratinolytic activity or keratinase enzyme are necessary [2]. Microbial degradation of the feather has attracted scientists and researchers as an affordable alternative approach to get feather powder with retained nutrients and essential amino acids. Keratinases are a special class of proteases that are able to hydrolyze keratin [3]. A number of microorganisms are capable to produce keratinase in the presence of keratin and can hydrolyze feather, hair, and nail and fur [8,13]. This enzyme is produced by a number of fungi and bacteria and numerous studies have led to identifying different strains of the bacterium Bacillus [9]. Since keratinases produced by fungi are known to be pathogenic, the use of bacterial keratinases is important [14]. The bacteria-based hydrolyzed keratin is used in the animal feed industry, nitrogen fertilizers, adhesives, films and also as a rich source serine, cysteine and proline [15,16]. In addition, keratinases are used in industries such as detergent industry, medicine, cosmetics and leather industry [17-19]. Agrahari and Wadhwa, isolated three strains of keratin-hydrolyzer bacteria, Bacillus megaterium strain SN2, Bacillus thuringiensis strains SN1 and Bacillus pumilus Strain SN3 from the soil of Gazipur area in India and all three isolates were able to hydrolyze chicken and penguin feathers at a certain time [6]. Zerdani and colleagues in Morocco, isolated 8 strains of Bacillus from the soil. Feathers in the medium were completely decomposed by all strains [5].
Materials and Methods
Collecting soil samples
The cross-sectional study was descriptive. Soil samples were collected from different poultry farms of Marvdasht, in the vicinity of Shiraz, in September 2015, from a depth of 5 to 10 cm, in sterile containers. All samples were collected in one day.
Isolation and screening keratinase producer bacteria
For each sample, 3 enrichment process (20-25 days) was done. In the first process, 5 gram of each soil sample was added to 100 ml of mineral salt medium in a 150 ml flask contained (grams per liter): NaCl 5, K2HPO4 1, KH2PO4 1, MgSO4 0.2, (NH4)2So4 1 and white feather 10, as the sole source of carbon and nitrogen, and the mixture was inoculated for 7 days at room temperature at Shaker 150 rpm. White feathers were thoroughly washed, then placed in boiling water for 10 minutes to separate wastes under the influence of heat [8,13]. To eliminate fats, feathers were placed in an equal mixture of chloroform: water (1:1) for 15 minutes [20]. Feathers then were rinsed several times with distilled water and were placed in the oven (100°C), for 2 hours to become completely dry [8,13].
In the second stage, 10 ml of the supernatant of the first enrichment was taken in sterile conditions and transferred to 90 ml of fresh liquid medium with 1% feather. All medium was stored at room temperature at Shaker 150 rpm. After 7 days, each sample was cultured on nutrient agar medium (Merck, Germany). This was repeated several times to obtain pure colonies [8,13].
The third stage was dedicated to the specific medium and only keratinase-contained bacteria were separated and identified. The enrichment was done on the Agar Based Mineral Salt Medium along with superfine feather (1 to 3 mm) as a source of carbon and nitrogen, the environment contained all salt-based medium ingredients and 15-gram agar. The purified colonies from nutrient agar medium were cultured on specific solid medium. Plates were incubated for 1 to 7 days at 37°C. Of these, five strains with higher feather hydrolyzing rate were collected for identification [8,13].
Identification by biochemical tests
Identification of bacteria was carried out by biochemical tests such as Gram staining, Sporulation staining, Mutilation test, Catalase test, Oxidase, Citrate, Starch hydrolysis [21-23].
Identification by 16S rRNA marker
To detect most bacteria by 16S rRNA, bacterial DNA was extracted by boiling. 1 cc of Brain Heart Infusion (Merck, Germany) medium containing pure bacteria, after 24 hours was centrifuged (Hettice, Germany) at 9300 g for 2 minutes. The supernatant was removed and 50 μl of sterile alcohol was added. The solution was shaken and boiled for 5 minutes, then centrifuged at 13400 g for 3 minutes. The supernatant was used as template DNA [9]. The 16s rRNA gene was amplified using Polymerase Chain Reaction (PCR) by universal primers:
• F (5- AGA GTT TGATCC TGG CTC AG-3)
• R (5-GGT TAC CTT GTTACG ACTT-3) [22]
The PCR solution contained 4 μl of DNA template, 0.6 μl of F primer, 0.6 μl of R primer, 8.2 μl of PCR Master mix (Taq DNA Polymerase Master Mix Red, Denmark), and 36.6 μl of distilled water in a final volume of 50 μl. PCR reaction was carried out according to the schedule, initial denaturation for 3 minutes at 94°C, denaturation for 1 minute at 94°C, primer annealing for 1 minute at 56°C, extension for 2 minutes at 72°C, final three stages were repeated 30 times, and the final step or the completion of synthesis was carried out for 10 minutes at 72°C (Techne, England) [24]. PCR products were sent for sequencing to the Korea Macro Research Laboratory. The 16S rRNA gene sequences were analyzed. Then, using the MEGA5.0 (Beta) software with Maximum likelihood, a phylogenetic tree was plotted for that strain with the highest keratin hydrolysis rate and sequences were registered in GenBank with an Accession number.
Measuring the activity of the keratinase enzyme
To measure enzyme activity, DMSO-Solubilized Keratin and Seed Culture are required.
Preparation of keratin-DMSO solution
Ten gram of the washed and degreased feathers was mixed with 500 ml of dimethyl sulfoxide solution (DMSO) and placed in a 100°C oven for 120 Min. Then, 1000 ml of cold acetone was added to the above solution and placed in a -24°C for 5 hours. From the solution, the feather precipitants were removed with centrifugation at 9300 g for 10 minutes, and then was rinsed thoroughly with distilled water three times and placed at 40°C for 2 hours until the precipitate was completely dried.
One gram of this sediment was dissolved in 20 ml of 0.05N NaOH. The pH of the solution was then adjusted to 8 using 0.1 mol/L Tris-HCl. The solution was used as a solution of keratin- DMSO [6].
Pure enzyme preparation (seed culture)
A number of highly feather hydrolyzer bacteria were cultured on nutrient agar for measurement of keratinase enzyme activity. After 24 hours, multiple colonies were taken from each sample and inoculated into liquid MSM medium with 1% feather as the sole source of carbon and nitrogen. Samples were stored at room temperature on a shaker at 150 rpm for 48 to 72 hours. 1 ml of supernatant of this medium was used as a pure enzyme [8,13].
Keratinase measurements
One ml of pure enzyme was mixed with 1 ml of keratin solution and placed in a water bath at 50°C for 10 minutes. This reaction was stopped after adding 2 ml of trichloroacetic acid (20% TCA). After centrifugation at 9300 g for 15 minutes, the optical absorbance of the supernatant was measured at 280 nm (A280) using a spectrophotometer (Cary50, Australia) in contrast to the control and was calculated by the following formula:
U= 4 × n × A280/(0.01 × 10)
0.01: The absorbance number indicated by the control, n: The dilution number, 4: The final volume in milliliters, 10: The duration of incubation in minutes.
By definition, one unit of enzyme activity is equal to the amount of enzyme that increases the optical absorbance 0.01 between the control and the specimen under the same conditions. The activity unit of the keratinase is reported in Units/ml/min. The culture medium with no substrate (feather) was used as the negative control [8,13].
Study of different conditions on the activity of a keratinase enzyme for superior strain and optimization of growth conditions:
Investigating the activity of keratinase enzymes at different temperatures: To obtain optimal temperature, the enzyme activity was measured at 25°C, 30°C, 35°C and 45°C [8,13].
Investigating the enzyme activity at different pH: To obtain the optimum pH of enzyme activity, the pH of the medium was adjusted to 6, 7, 8, 9 and10 [8,13].
Investigating the enzyme activity at different concentrations of substrate (feather): To obtain an optimal concentration of substrate, the bacterium was cultivated in different percentages of 0.25%, 0.5%, 0.75%, 1.5% and 2% of a feather [8,13].
Investigating the activity of the enzyme in different concentrations of glucose as an additional source of carbon: To obtain optimal enzyme activity, the bacterium was cultured in different percentages (2.5%, 1.25%, 0.5%) of glucose and the enzyme activity was measured for 5 consecutive days [8,13].
All of the above experiments were performed three times, and enzymatic activity was measured for 5 days in a superior strain.
Data analysis
After measuring the enzyme activity, variables were analyzed using SPSS software and ANOVA test at the significance level of 0.01 (p<0.01).
Results
Of the total soil samples analyzed, 30 different species of bacteria were isolated on nutrient agar medium. To isolate keratinase producer bacteria, 30 isolates were transferred to the feather-rich medium at 37°C and 15 isolates used the feather as the sole carbon source. Five isolates showed good hydrolysis power and were selected for identification.
Five bacterial strains were identified through microscopic examination and biochemical tests. Results are shown in Tables 1 and 2.
| SKH1 | SKH2 | SKH3 | SKH4 | SKH5 | |
|---|---|---|---|---|---|
| Gram reaction | positive | positive | positive | positive | positive |
| Spore forming | central | terminal | central | Sub terminal | terminal |
| Morphological-microscopic | Bacillus (single or streptobacillus) | Bacillus | coccobacillus | Bacillus | Bacillus |
| Need for oxygen | facultative-aerobic | aerobic | Facultative-aerobic | aerobic | aerobic |
| Colony color | Yellowish creamy | white | creamy | Creamy white | creamy |
| Colony shape | Convex-smooth-small | Smooth-rough-large | Convex-smooth-large | Smooth-rough-large | Smooth-rough-large |
Table 1: Morphological-microscopic features of 5 selective bacteria.
| SKH1 | SKH2 | SKH3 | SKH4 | SKH5 | |
|---|---|---|---|---|---|
| Catalase | Positive | Positive | Positive | Positive | Positive |
| Oxidase | Negative | Negative | Negative | Negative | Negative |
| Citrate | Positive | Negative | Positive | Negative | Negative |
| Starch Hydrolysis | Positive | Positive | Positive | Positive | Positive |
| VP test | Positive | Negative | Positive | Negative | Negative |
| Methyl red test | Negative | Negative | Negative | Negative | Negative |
| Nitrate Fermentation | Positive | Positive | Positive | Positive | Positive |
| Carbohydrate Fermentation | A/- | -/- | A/- | -/- | -/- |
Table 2: Some bio-chemical features of 5 selective bacteria.
Based on Gram staining and some biochemical tests, it was demonstrated that one isolated strain is Bacillus cereus, one is Bacillus Thuringiensis, and three isolates are Bacillus flexus.
The results of PCR product
After DNA extraction and PCR, PCR products obtained for each bacteria were analyzed on 1.5% agarose gel and after visualization by Gel Doc (BioRAD, USA) it was observed that 16S rRNA genes had created 1500 bp bands (Figure 1). The results of sequencing were registered in the Gene Bank, as a new strain (Table 3). 16s rRNA sequence and Phylogenetic tree (Figure 2), indicating that Bacillus cereus SKH1 is phylogenetically close to the Bacteria Bacillus cereus G12.
| Bacterium name | strain | Specific number |
|---|---|---|
| Bacillus cereus | SKH1 | KX371092 |
| Bacillus flexus | SKH2 | KX343199 |
| Bacillus thuringiensis | SKH3 | KX371093 |
| Bacillus flexus | SKH4 | KX371094 |
| Bacillus flexus | SKH5 | KX371095 |
Table 3: Bacterium name, strain, and the specific name registered in the Gene Bank.
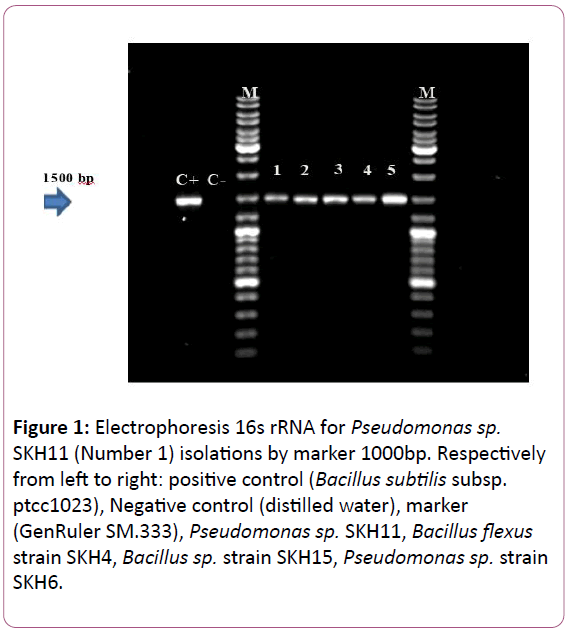
Figure 1: Electrophoresis 16s rRNA for Pseudomonas sp. SKH11 (Number 1) isolations by marker 1000bp. Respectively from left to right: positive control (Bacillus subtilis subsp. ptcc1023), Negative control (distilled water), marker (GenRuler SM.333), Pseudomonas sp. SKH11, Bacillus flexus strain SKH4, Bacillus sp. strain SKH15, Pseudomonas sp. strain SKH6.
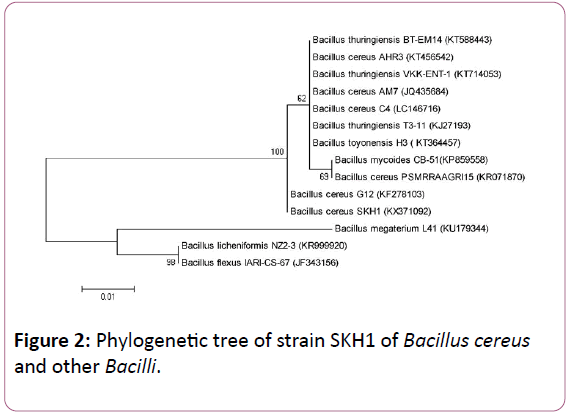
Figure 2: Phylogenetic tree of strain SKH1 of Bacillus cereus and other Bacilli.
Keratinase enzymatic activity measurement
All five selected strains were transferred the feathercontained MSM liquid medium (Figure 3). Full feather hydrolysis time, as well as the number of base pairs, registered gene are shown in Table 4. Feathers were fully hydrolyzed from 72 hours to 100 hours for 5 different strains.

Figure 3: Full decomposition of feathers by Pseudomonas sp. strain SKH11 after 24 hours (left Erlenmeyer flask), (right Erlenmeyer flask, growth medium before bacterium inoculation).
| Bacterium name | Number of base pairs | Duration of full decomposition of feathers |
|---|---|---|
| B. cereus SKH1 | 508bp | 72 hours |
| B. flexus SKH2 | 529 bp | 100 hours |
| B.thuringiensis SKH3 | 500 bp | 72 hours |
| B. flexus SKH4 | 850 bp | 90 hours |
| B. flexus SKH5 | 1061 bp | 100 hours |
Table 4: Duration of full decomposition of feathers, full names of bacteria, and a number of base pair.
Enzyme activity measurement
Keratinase activity was measured for the 5 mentioned bacterial strains, compared with positive control samples (Bacillus subtilis subsp. PTCC1023) based on a unit per milliliter per minute (Unit/ml/min), respectively. The values obtained are measured only once. The highest activity was related to Bacillus cereus SKH1, while the lowest enzyme activity was related to Bacillus flexus SKH4. The results are shown in Table 5.
| Bacterium name | Enzyme activity (Uint/ml/minute) |
|---|---|
| Bacillus cereus SKH1 | 17.12 |
| Bacillus flexus SKH2 | 13.26 |
| Bacillus thuringiensis SKH3 | 15.56 |
| Bacillus flexus SKH4 | 10.81 |
| Bacillus flexus SKH5 | 11.95 |
Table 5: Amount of enzyme activity of different bacterial strains SKH1- SKH5.
Measurement of the keratinases enzyme activity under different conditions
Since Bacillus cereus SKH1 showed the most analysis activity in less time with the best enzymatic activity, it was selected for the study of enzyme activity in different conditions. Analyzed by ANOVA test in three periods with changing conditions led to the following results.
Comparing the effect of temperature on keratinase production: The overall result of the study of Bacillus cereus SKH1 enzyme activity in the temperature range of 25°C, 30°C, 35°C, 40°C and 45°C on different days shows that at 25°C, 30°C, 35°C, 40°C and 45°C, the bacterium is able to grow and produce the enzyme. The optimal temperature to maximize the production of the enzyme was 35°C. At 25°C, the minimum enzyme activity was observed and at temperatures of 40°C and 45°C, the activity was mild (Figure 4).
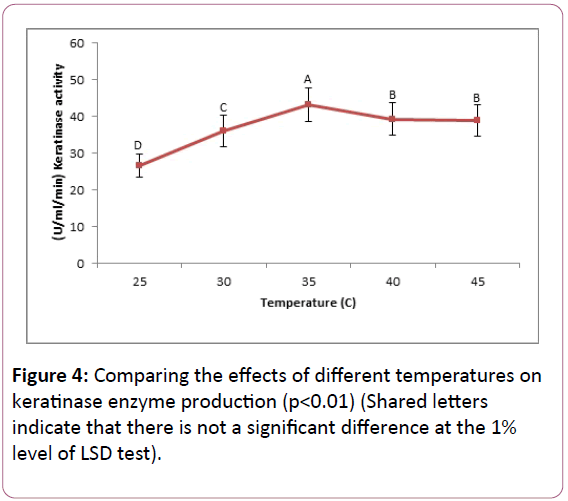
Figure 4: Comparing the effects of different temperatures on keratinase enzyme production (p<0.01) (Shared letters indicate that there is not a significant difference at the 1% level of LSD test).
Comparing the effect of initial pH on keratinase production: Bacillus cereus SKH1 keratinase activity measurements showed that the bacterium grows and produce keratinase in the pH range of 6, 7, 8, 9 and 10. The optimum pH for maximum production of the enzyme was 8. The bacterium produces the lowest enzyme at neutral pH 6 and in pH 9, showed mild enzymatic activity (Figure 5).
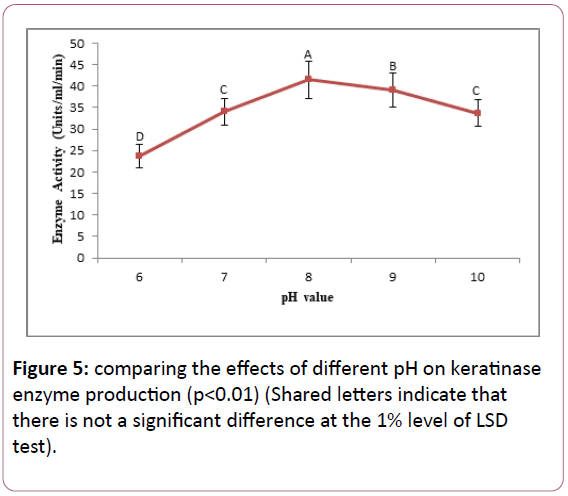
Figure 5: comparing the effects of different pH on keratinase enzyme production (p<0.01) (Shared letters indicate that there is not a significant difference at the 1% level of LSD test).
Comparing the effects of different amounts of substrate (feather) on the production of keratinase: Result showed that Bacillus cereus SKH1 is able to grow and produce keratinase in different substrate amounts: 0.25%, 0.5%, 0.75%, 1.5%, and 2%. The optimal value of substrate, for maximum enzyme production, was determined 0.75%. The bacterium showed the lowest enzyme production in the amounts of 0.25% and 2% of the substrate. The two values were not significantly different based on the LSD test with 1% significance level (Figure 6).
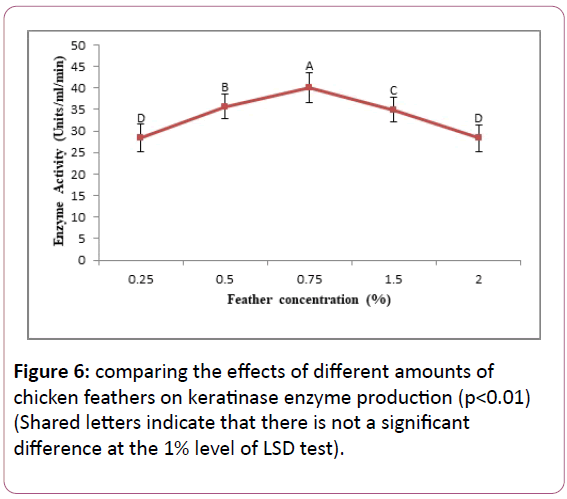
Figure 6: comparing the effects of different amounts of chicken feathers on keratinase enzyme production (p<0.01) (Shared letters indicate that there is not a significant difference at the 1% level of LSD test).
Comparing the effects of different amounts of glucose on the enzyme production: The results showed that the strain SKH1 is able to grow and produce keratinase with or without glucose and also in different levels of this simple sugar.
The bacterium in the absence of glucose shows the maximum keratinase activity. In the presence of glucose, the most keratinase activity was related to the glucose concentration of 1% (Figure 7).
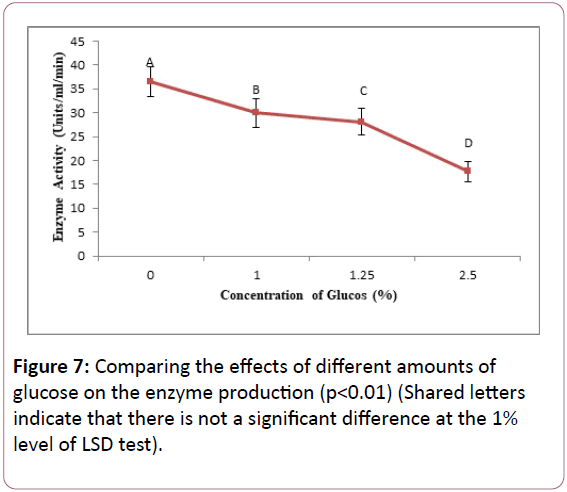
Figure 7: Comparing the effects of different amounts of glucose on the enzyme production (p<0.01) (Shared letters indicate that there is not a significant difference at the 1% level of LSD test).
Discussion
Bacillus species are one of the best keratinase producers with higher capacity production of this enzyme, among other producers (about 20 to 25 grams per liter) [15]. A number of researchers, like Femi-Ola and colleagues, Deivasigamani and his colleagues, Mehta, and colleagues isolated several species of Bacillus from the soil or poultry waste with the ability to produce keratinases [8,13,21,22]. In the present study, from Marvdasht, 15 keratinase producer bacteria were isolated, 5 isolates with the more power to produce keratinases were selected for further evaluation and after identification by morphology, biochemical tests and 16S rRNA sequencing, three different species of the genus Bacillus called Bacillus cereus, Bacillus thuringiensis and Bacillus flexus, were identified. For each of these five strains, the specific number was determined in GenBank. Femi-Ola et al. and Mazotto et al., isolated keratin hydrolyzer Bacillus cereus from soil [18,19,22]. Sivakumar et al., and Agrahari et al., similar to this study, isolated Bacillus thuringiensis from soil [6,25]. So far, in Iran, no one has isolated keratinase producer Bacillus flexus from the soil. Five samples bacterial enzyme activity was evaluated in one phase to identify the strongest strains. All strains showed good enzymatic activity and Bacillus cereus SKH1 was determined as the strongest strain (Table 4) and this was similar to Mehta and colleagues, which determined the enzymatic activity of B. licheniformis 14.344 U/ml/min, and Agrawal et al. that determined the enzymatic activity of a genus of Bacillus 13.60 U/ml/min [8,10,13].
The goal of optimization is to find conditions affecting the bacterial enzyme production. Temperature is another physical parameter affecting the production of enzymes. Temperature controls the synthesis of enzymes and energy metabolism [8,13].
Bacillus cereus SKH1 showed varying amounts of enzyme release in the range of 25°C, 30°C, 35°C, 40°C, and 45°C. The optimum activity for the enzyme production in this bacterium was determined 35°C. The bacterium is thermophile and grows at high temperature and needs less time for a full hydrolysis. The results of this study were in line with the results of Agrawal et al., determined the optimum temperature for Bacillus strain equal to 37°C and Mehta and colleagues determined the optimum temperature for Bacillus subtilis equal to 35°C [8,10,13]. Bacillus species are able to grow in the temperature range of 30°C to 55°C. Although in some cases, keratinolytic bacteria are also able to produce keratinase at higher temperatures [8,13]. Some studies have reported the temperature range of 40°C to 60°C [24]. Park et al., for Bacillus megaterium and Infante et al., for Bacillus thuringiensis, reported the optimum temperature for maximum production keratinase equal to 40°C and Vigneshwaran et al., for Bacillus licheniformis, reported the optimum temperature of 60°C [24].
The initial pH of culture media affects fermentation processes and the transmission of nutrients across the cell wall. Enzymes are active within a certain pH. Cell wall enzymatic activity is a set of functions related to the initial pH [8,13]. Optimum pH to produce the greatest amount of keratinases by bacteria was 8 and was in accordance with previous reports by Talebi et al., for Bacillus pumilus and Deivasigamani and his colleagues for a strain of Bacillus. Riffel et al. also found similar results with the most activity in the pH of 7.5. Park and his colleagues determined the best pH for one strain of Bacillus equal to 7.4 which is similar to our results [21,26-28]. A number of studies have reported different pH for optimum production of keratinases. Infante and colleagues for Bacillus thuringiensis, Sivakumar for Bacillus thuringiensis and Mehta and colleagues for Bacillus Sonorensis reported the best pH equal to 7, 10 and 7, respectively. Mousavi and colleagues reported the optimum pH for Bacillus subtilis keratinases equal to 11 that is different from the results obtained in this study [8,9,13,25,29]. It is important to note that all of these reports indicate that the activity of bacterial keratinases is optimum in the range of alkaline pH.
It is expected that keratinase producer bacteria use feather keratin for growth and proliferation. The maximum amount of substrate for optimum keratinase production by SKH1 strain was about 0.75. This is in line with the study of Mehta et al. [8,13] but is inconsistent with Pandian et al. who reported that about 2% for some strains of Bacillus [2]. Because production of keratinase is dependent on the presence of keratin, increasing the substrate saturate the substrate (feather), which is, in turn, reduces the quality of keratinase, and reduce the optimum activity [20]. As a result, increasing amounts of substrate (2% WV of a feather), enzymatic activity will be reduced. Also, at low amounts of substrate (0.25 percent) due to lack of available keratin to bacteria as the sole source of carbon and nitrogen, enzymatic activity will be reduced as well.
It was also observed that in the presence of excess glucose, hydrolyzes of feathers reduced subsequently compared to medium without glucose. In the absence of excess carbon sources, feathers were used as the sole carbon source and enzyme production was increased [8,13]. But adding carbon source (glucose) prevents the secretion of keratinase. The presence of simple carbon sources in addition to a source of rigid proteins such as keratin reduces enzyme production. Bacteria use simple food supplies at first [8,13]. In this case, the bacteria use the medium glucose as a carbon source. In this research, as expected, Bacillus cereus SKH1 showed its maximum enzyme production in the medium with a feather as the only carbon source. This is consistent with the findings of Mehta and colleagues for the bacteria Bacillus sonorensis, and the findings of Sivakumar and his colleagues for Bacillus thuringiensis [8,13,25].
Conclusion
The hydrolysis of a feather by Bacillus cereus SKH1 confirms that this bacterium has a substantial protease activity and can break keratin disulfide bonds. Measuring enzymatic activity in Five isolated strains showed that SKH1 strain may be the best candidate in various industries.