Cordula Enenkel*
Department of Biochemistry, University of Toronto, University Avenue, Toronto, Ontario, Canada
Corresponding Author:
Cordula Enenkel
Department of Biochemistry University of Toronto University Avenue, Toronto, Canada.
Tel: 001 416 978 3843
Fax: 001 416 978 8545
E-mail: cordula.enenkel@utoronto.ca
Received Date: April 11, 2017; Accepted Date: May 16, 2017; Published Date: May 19, 2017
Citation: Enenkel C. Proteasome Dynamics: Will the Territory of Proteasomes Be Claimed By Mitochondrial Proteases Under Stress Conditions? Biochem Mol Biol J. 2017, 3:2. doi: 10.21767/2471-8084.100036
Short Communication
Proteasomes are responsible for the ubiquitin-dependent degradation of proteins during cell proliferation. Their 26S holo-enzymes are composed of multiple subunits arranged in a proteolytic core particle and a regulatory particle which recognizes the substrates by covalently attached poly-ubiquitin chains. Proteasomal substrates are short-lived and if not eliminated are prone to aggregation [1]. Among these substrates are improperly folded nascent polypeptides as well as many other proteins regulating cellular processes such as cell cycle progression [2]. In yeast, about thirty strains are available expressing individual proteasomal subunits as fusions with the green fluorescent protein (GFP) [3]. Each GFP fusion protein replaces the endogenous subunit, thus is a suitable reporter on proteasome localizations. The scientific community was surprised by the results of subsequent in vivo localizations that revealed proteasomes to primarily localize to the nucleus of yeast cells grown to logarithmic phase. Transiently expressed GFPtagged versions of mammalian subunits which are adequately incorporated into the proteasome showed a major nuclear localization as well [4] confirming early proteasome localizations in cancer cell lines [5]. The question arose as to where misfolded and aggregation-prone proteins might be degraded. If proteasomes are mainly nuclear, would enough proteasomes be present in the cytoplasm for the degradation of misfolded cytoplasmic proteins? Certainly, the quality and not the quantity of proteasomes will account for proteolytic activity. Kiran Madura stated that proteasomes are hardly active in the nucleus [6], while Yasushi Saeki and his co-workers reported that yeast cells proliferate even if cytosolic proteasomes are diminished [7]. Intriguingly, proteasomes can be imported as inactive precursor complexes into the nucleus, leaving uncertainty to which extent nuclear proteasomes are proteolytically active [8]. Late steps of their maturation [9] may take place at the nucleoplasmic basket of the nuclear pore, where Esc1, Ecm29 and Sts1/Cut8 coordinate the assembly and anchorage of nuclear proteasomes [10]. In parallel, nuclear import of active holo-enzymes exists as well [11] and prevails when inactive precursor complexes are not yet available upon exit from quiescence with the resumption of cell proliferation [12].
The intracellular dynamics of proteasome substrates are even more confusing than the movements of proteasomes between the nucleo- and cytoplasm. The dynamics of proteasomes and their substrates can be thoroughly studied in yeast, since yeast division occurs through closed mitosis, where the nuclear envelope remains intact. Evidence is found in the literature that nuclear substrates exit the nucleus and cytoplasmic substrates enter the nucleus for proteasomal degradation [13-15]. The polyubiquitination of misfolded cytosolic proteins is mainly achieved by two ubiquitin ligases, the cytoplasmic Ubr1 ligase and the nuclear San1 ligase. For poly-ubiquitylation through San1 a cytoplasmic protein has to be imported into the nucleus. Also the size of the substrate seems to determine where it is degraded. Cytosolic proteins with a molecular mass of around 60 kDa, the diffusion barrier of the nuclear pore complex, are susceptible for San1-mediated proteasomal degradation, while higher molecular mass proteins remain in the cytoplasm [16].
Growth under stress either by heat shock or nutrient deprivation triggers the reorganization of multiple cellular components [17]. Upon heat shock many proteins are sequestered into stress granules, though proteasomes are not found within stress granules [18]. When glucose is depleted and the ATP level is decreased, yeast cells arrest cell cycle progression and enter quiescence, alternatively named stationary or G0 phase. With the transition from proliferation to quiescence proteasomes exit the nucleus and accumulate in motile and reversible cytoplasmic storage granules [19]. Almost nothing is known about the proteolytic function of proteasome storage granules. At least aberrant nascent polypeptides will rarely emerge as potential substrates, since quiescent cells reduce the synthesis of new proteins [17].
A recently published work by Rong Li and co-workers revealed a fascinating protein degradation pathway named MAGIC (mitochondria as guardian in cytosol). Under heat stress conditions mitochondrial proteases were found to be involved in the degradation of cytosolic proteins prone to aggregation, when proteasomes are mainly sequestered into granules [20]. The inhibition of proteasomes had only a minor effect on the degradation of these substrates, while blocking mitochondrial import resulted in the aggregation of cytoplasmic substrates. Also the timely dissolution of cytosolic protein aggregates required mitochondrial import and proteases [21].
Mitochondrial import of nuclearly encoded proteins which are synthesized by ribosomes in the cytoplasm usually contain N-terminal presequences, which form in the canonical sense a net positively charged amphipathic helix [22]. The general entry gate for precursor proteins into the mitochondria is the TOM translocase in the outer mitochondrial membrane. To bridge the intermembrane space between the outer and inner mitochondrial membrane the precursor protein is handed over to the TIM translocase embedded in the inner mitochondrial membrane, which allows the translocation of the precursor protein into the mitochondrial matrix. Heat shock proteins assist the unfolding and refolding of the translocating precursor protein. The N-terminal presequence is cleaved off by the mitochondrial processing protease, once it arrives in the matrix [23].
Both nuclearly and mitochondrially encoded proteins form the mitochondrial proteome which is monitored by a protein quality control system. Especially under stress conditions and upon the formation of reactive oxygen species, aberrant proteins arise at the inner mitochondrial membrane and within the mitochondrial matrix. To maintain mitochondrial functions damaged proteins must be degraded. The mitochondrial protein degradation machinery relies on i-AAA, m-AAA, LON protease and the mitochondrial ortholog of ClpX/ClpP (for review see [24]. These proteases require ATP to degrade unfolded substrates. Since ATP is generated within mitochondria, it is not limiting under healthy conditions. However, during aging mitochondria dysfunctions accumulate and cause a vicious cycle of disturbed protein homeostasis linked with the onset of neurodegenerative diseases [25].
With regard to mitochondrial import of aggregation-prone cytoplasmic proteins, it remains magic how these proteins pass the outer and inner mitochondrial membrane for degradation. Though mitochondrial proteases claim the territory which proteasomes monopolized so far in the degradation of these cytoplasmic proteins, particularly under stress conditions, the mechanism of the mitochondrial uptake of aberrant cytosolic proteins remains elusive. In the nucleus however, proteasomes and San1 ubiquitin ligase appear to be abundant and able to achieve the degradation of aggregation-prone nuclear proteins even under stress conditions [26,27].
The model depicts a section into a yeast cell showing the dynamics of proteasomes upon the transition from proliferation to quiescence. On the upper left corner a proteasome storage granule is shown as a densely packed membrane-less spherical organelle within the cytoplasm. On the lower right corner the nucleoplasm is shown with barrel shaped proteasome holoenzymes. One proteasome is in the process of passing the nuclear basket of the nuclear pore to exit the nucleoplasm. Few proteasomes are stuck in the central plug of the nuclear pore, while others already arrived at the cytoplasmic filaments of the nuclear pore and migrate towards the proteasome storage granule. Nuclear pores are depicted as craters embedded into the double membrane of the nuclear envelope. For simplification only proteasomes are shown. In reality the nucleo- and cytoplasm is crowded with proteins (Figure 1).
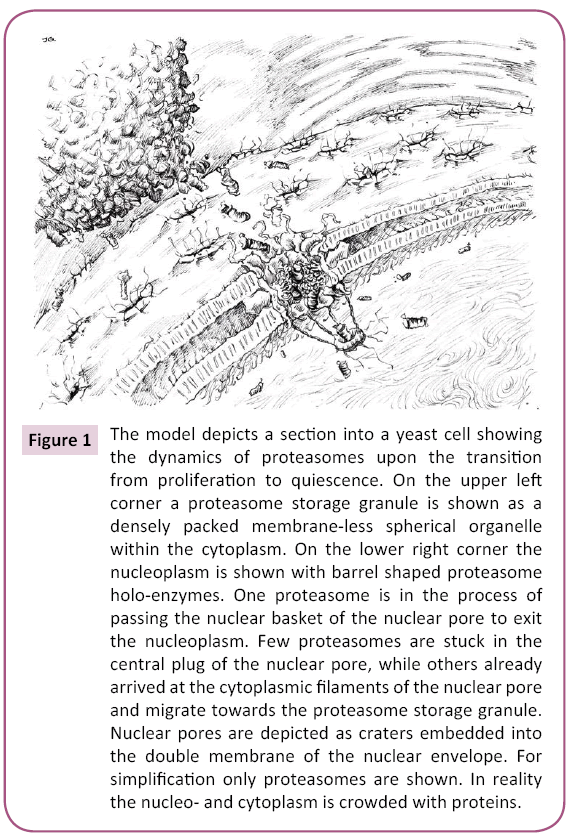
Figure 1: The model depicts a section into a yeast cell showing the dynamics of proteasomes upon the transition from proliferation to quiescence. On the upper left corner a proteasome storage granule is shown as a densely packed membrane-less spherical organelle within the cytoplasm. On the lower right corner the nucleoplasm is shown with barrel shaped proteasome holo-enzymes. One proteasome is in the process of passing the nuclear basket of the nuclear pore to exit the nucleoplasm. Few proteasomes are stuck in the central plug of the nuclear pore, while others already arrived at the cytoplasmic filaments of the nuclear pore and migrate towards the proteasome storage granule. Nuclear pores are depicted as craters embedded into the double membrane of the nuclear envelope. For simplification only proteasomes are shown. In reality the nucleo- and cytoplasm is crowded with proteins.
Acknowledgement
C.E. is supported by NSERC (4422666-2011) and CIHR (325477). Thanks to Zhu Chao (Jerry) Gu for drawing the image of proteasome storage granules in the cytoplasm.
References
- Amm I, Wolf DH(2016) Molecular mass as a determinant for nuclear San1-dependent targeting of misfolded cytosolic proteins to proteasomal degradation. FEBS Lett 590: 1765-1775.
- Amsterdam A, Pitzer F, Baumeister W(1993) Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc Natl Acad Sci USA. 90: 99-103.
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N(2009) Importing mitochondrial proteins: machineries and mechanisms. Cell138: 628-644.
- Chen L, Madura K(2014a) Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics 197: 193-197.
- Chen L, Madura K(2014b) Yeast importin-alpha (Srp1) performs distinct roles in the import of nuclear proteins and in targeting proteasomes to the nucleus. J Biol Chem 289: 32339-32352.
- Enenkel C(2014) Proteasome dynamics. Biochim Biophys Acta 1843: 39-46.
- Hamon MP, Bulteau AL, Friguet B(2015) Mitochondrial proteases and protein quality control in ageing and longevity. Ageing Res Rev 23: 56-66.
- Hershko A, Ciechanover A, Varshavsky A(2000) Basic Medical Research Award: The ubiquitin system. Nat Med 6: 1073-1081.
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, et al.(2016) UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell 166: 935-949.
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425: 686-691.
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, et al.(2016) ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164: 487-498.
- Laporte D, Salin B, Daignan-Fornier B, Sagot I(2008) Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181: 737-745.
- Lehmann A, Janek K, Braun B, Kloetzel PM, Enenkel C(2002) 20S proteasomes are imported as precursor complexes into the nucleus of yeast. J Mol Biol 317: 401-413.
- Lehmann A, Niewienda A, Jechow K, Janek K, Enenkel C(2010) Ecm29 fulfils quality control functions in proteasome assembly. Mol Cell 38: 879-888.
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, et al. (2012) Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671-683.
- Medicherla B, Goldberg AL(2008) Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol 182: 663-673.
- Miller SB, Mogk A, Bukau B(2015) Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol 427: 1564-1574.
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, et al.(2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell 24: 3920-3938.
- Pack CG, Yukii H, Tohe A, Kudo T, Tsuchiya A(2014) Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun 5: 3396.
- Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, et al. (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154: 134-145.
- Prasad R,KawaguchiS, Ng DT(2010) A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell 21: 2117-2127.
- Ruan LC, Zhou E, Jin A, Kucharavy Y, Zhang Z, et al. (2017) Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543: 443-446.
- Schulz C, Schendzielorz A, Rehling P(2015) Unlocking the presequence import pathway. Trends Cell Biol 25: 265-275.
- Takeda K, Tonthat NK, Glover T, Xu W, Koonin EV, et al.(2011) Implications for proteasome nuclear localization revealed by the structure of the nuclear proteasome tether protein Cut8. Proc Natl Acad Sci USA. 108: 16950-16955.
- Tsuchiya H, Arai N, Tanaka K, Saeki Y(2013) Cytoplasmic proteasomes are not indispensable for cell growth in Saccharomyces cerevisiae. Biochem Biophys Res Commun436: 372-376.
- Voos W, Jaworek W, Wilkening A, Bruderek M(2016) Protein quality control at the mitochondrion. Essays Biochem 60: 213-225.
- Weberruss MH, Savulescu AF, Jando J, Bissinger T, Harel A, et al.(2013) Blm10 facilitates nuclear import of proteasome core particles. EMBO J 32: 2697-2707.


