Research Article - (2022) Volume 8, Issue 1
Photosystem Proteins Protects by Small Heat Shock Protein in Cyanobacteria under Oxidative Stress Condition
Kollimalai Sakthivel*
Department of Marine Biology, Annamalai University, India
*Correspondence:
Kollimalai Sakthivel, Department of Marine Biology, Annamalai University,
India,
Email:
Received: 03-Jan-2022, Manuscript No. IPBMBJ-22-11703;
Editor assigned: 05-Jan-2022, Pre QC No. IPBMBJ-22-11703 (PQ);
Reviewed: 19-Jan-2022, QC No. IPBMBJ-22-11703;
Revised: 25-Jan-2022, Manuscript No. IPBMBJ-22-11703 (R);
Published:
31-Jan-2022, DOI: 10.36648/2471-8084.22.8.55
Abstract
Chaperone activity of small heat shock proteins HSP with Photosystem I Protein (PsaAB) in the presence of hydrogen
peroxide was studied. HspA plays a protective role under oxidative stress in Synechococcus elongatus strain
ECT16-1, which constitutively expresses HspA. Reference strain ECT, which does not constitutively express HspA,
ECT16-1 showed much better chlorophyll in hydrogen peroxide. Photosystem protein PsaAB was degraded on 72
hours incubation in ECT but there was no change in ECT16-1. The PsaAB absorbance reduced at 65% on 0.3% hydrogen
peroxide at 30°C for 9 hours incubation. Two degradation product of photo system protein PsaAB (40, 13
kDa) were detected and those bands intensity was increased at higher concentration of hydrogen peroxide. The
PsaB one of the major photo system I protein degradation product were also detected by western blott analysis.
The degradation of photo system I protein (PsaAB) was observed at the low concentration of hydrogen peroxide
(0.15%) in control, and lysozyme mixed samples, α-crystallin is the homologus protein of small heat shock protein
protected up to 0.3% and HspA protected at high concentration of hydrogen peroxide on 0.4%. In western Blotting
analysis showed the photo system I protein (PsaB) not degraded when the sample mixed with α-crystallin in the
presence of 0.3% hydrogen peroxide. Spectrophotometer results shows when photo system I peak was reduced
due to the hydrogen peroxide mixed with buffer (control-1) or lysozyme (control-2), but the peak not reduces in
HspA mixed photo system I.
Introduction
It was suggested that the accumulation of heat shock protein
is correlated with thermos tolerance [1]. Subsequently it has
been recognized that HSPs have a protective role against a variety
of stresses beside high temperature. These include nitrogen
starvation [2], hyperosmotic stress, and salt stress [3,4],
oxidative stress [5,6]. Oxidative species like superoxide (O2),
hydrogen peroxide (H2O2) and hydroxyl radicals (OH) are generated
as a result of incomplete reduction of oxygen during respiration
and photosynthesis. They can damage the cell, and it
is conceivable that DNA repair, protein degradation, metabolic energy generation, cell division, and other cellular activities are
all coordinately regulated [7,8].
Chloroplasts have arisen from a cyanobacterial ancestor [9]. It
is maintained an independent circular genome encoding gene
products which are important for primary and secondary processes
of photosynthesis. Photosystem I (PSI) is a multisubunit
pigment-protein complex of oxygenic photosynthesis located
in the thylakoid membrane of chloroplasts and cyano bacteria.
The PSI subunits are designated psaA-P, according to the corresponding
psaA-P genes [10,11]. A heterodimer of two integral
membrane proteins, PsaA and PsaB, forms the core of the PSI
reaction centre, which binds the electron transfer components P700, Ao, A1, and Fx. It has been reported that adding the CP
sHSP 21, to thylakoid membranes in vitro protects against heat
inactivation [12]. Photoinhibition in chilling-sensitive plants did
occur during chilling stress under low irradiance [13]. It was reported
that the activity of PSI decreased about 70-80%, when
cucumber leaves were treated at 5ºC in an irradiance of 100
μmol m-2s-1 In Synechocystis sp. PCC 6803, PSI genes are actively
transcribed under low-light (LL) conditions, whereas their
transcription is coordinately and rapidly down-regulated upon
shift to HL conditions [14-17]. PsaA and PsaB proteins, the heterodimer
subunits of the PSI reaction centre, were degraded by
photo inhibition at chilling temperatures [18].
For the present study we purified photosystem I proteins from Synechococcus sp. PCC 7942 in order to analyze the role of heat
shock proteins in preventing degradation and aggregation under
oxidative stress conditions. Hydrogen peroxide is one of
the oxidative species that were used for creating the oxidative
stress condition artificially. We found that HspA one of the small
heat shock proteins, protect and photo system I, from degradation
and aggregation during oxidative stress.
Materials and Methods
Organisms and Culture Conditions
The cyanobacterial strain Synechococcus sp. PCC 7942 was cultured
photoautotrophically in BG-11 inorganic liquid medium
or on BG-11 plates containing 1.5% (w/v) agar and 0.3% (w/v)
sodium thiosulphate. The BG-11 culture medium was modified
with 50 μg/ml Na 2CO3 and 5 mM 2-[4-(2-Hydroxyethyl)-1-piperazinyl]
ethanesulfonic acid and adjusted to pH 8.0 with KOH.
The liquid cultures in glass vessels were incubated at 30°C, continuously
aerated and illuminated with a light intensity of 30
μmol photons m-2 s-1.
Preparation of Thylakoid Membrane
Cultures of Synechococcus sp. PCC 7942 in the logarithmic
growth phase (0.7 OD at 730 nm) were harvested by centrifugation
at 3000 rpm for 15 min. The pellet was resuspended
in ten volumes of thylakoid buffer 25 mM MES/NaOH, pH 7.0
containing 1 mM aminocaproic acid. The cells were disrupted
using anequal amount of glass beads (0.177-0.25 mm in diameter)
and vortexed twice for 1 min with 2 min interruption for
cooling on ice. Glass beads were washed four times with 200
μl of thylakoid buffer, aliquots pooled and unbroken cells were
removed by centrifugation at 1000 rpm for 5 min. The supernatant
containing thylakoid membranes was centrifuged at 13,000
rpm for 30 min. Thylakoid membranes were re suspended in
thylakoid buffer to a concentration of about 500 μg Chl ml-1.
Isolated membranes were highly enriched with thylakoid and
stored at -20°C.
Purification of Photosystem Proteins
The thylakoid membranes were diluted in thylakoid buffer to a
chlorophyll concentration of 250 μg ml-1 and solubilized with
Triton ×100 (2% final concentration) for 15 min onice. Nonsolubilized
material was removed by centrifugation at 13,000
rpm for 30 min at 4°C. Then the supernatant was centrifuged at 70,000 rpm (HITACHI Himac CS 120 FX, S120AT2-0157 rotor) for
60 min at 4°C. Photosystem II particles were solubilized in the
supernatant, while the sediment was enriched in Photosystem
I.
Purification of Small Heat Shock Proteins
Small heat shock protein purified from HspA gene amplified Synechococcus sp. PCC 7942 chromosomal DNA with two oligonucleotide
primers, HspA-F(5-ATATGGCACTCGTTGATTC-3) and
HspA-R(5- CTCGAGTCGCTCGCAAGCTTCA G-3) by a previously
described procedure. The amplified fragment was cloned into
pT7Blue Vector (Invitrogen) according to the manufacturer’s instructions.
This plasmid was digested with NdeI and XhoI and
the insert was subsequently cloned into the pET21a expression
vector (Invitrogen), previously digested with the same restriction
enzymes. The vector pET21a-HspA carried a His6 tag
which was fused to the C terminus of the hspA gene product.
The constructed plasmid was transformed into E.coli BL21(DE3).
The cells were grown to apparent absorption 0.6 at 540 nm in
LB medium containing 100 μg/ml ampicillin at 37°C, and then
IPTG was added to a concentration of 1 mM. After a further 3 h
at 37°C, the cells were harvested by centrifugation and stored
at -80°C. The pellet was suspended TE buffer (50 mMTris/HCl,
pH 7.5 and 1 mM EDTA) and lysed by sonification. After removing
cell debris the protein in the supernatant was precipitated
with 0-40% (w/v) saturated ammonium sulphate. The protein
pellet was dissolved in TE buffer and loaded onto DEAE/Toyopearl
650S equilibrated with TE. After washing the column with
TE, HspA was eluted with a 0-250 mM NaCl gradient in TE and
fractionated. Fractions were examined by SDS/PAGE to identify
those containing HspA. Fractions containing HspA were pooled,
dialyzed against TE and applied to a column of hydroxylapatite
HP40-100 equilibrated with TE. After washing the column with
TE, protein was eluted with a 0-400 mM Na-phosphate (pH
7.7) gradient. Fractions containing HspA were pooled, dialyzed
against TE and stored at -20°C. α-crystallin source from Bovine
Eye Lens, purchased from Stressgen Bioreagent Corp. and molecular
weight 20.037 kDa.
Stress Condition
Photosystem protein was mixed with heat shock proteins (1:1),
in the presence of 0.05 to 7% hydrogen peroxide and incubated
at 30˚C for 3 hours for SDS-PAGE, Western Blotting, absorption
and fluorescence spectrophotometry. For characterization
studies, photosystem protein and hydrogen peroxide at 0.03-
0.3%, temperature 0 and 37˚C incubated for 9 hours and optimum
density at 663 nm was measured by a Shimadzu UV-1200
double beam spectrophotometer (Shimadzu, Kyoto, Japan).
BN/SDS-PAGE and Western Blot Analysis
Thylakoid membrane (14 μg Chl.) was solubilized with 2% Triton
X-100 and loaded on 7% BN-PAGE at 4˚C. Soluble fraction
(40 μg protein) was mixed with equal amount of SDS-sample
buffer containing 0.12 M Tris/HCl (pH 6.8), 0.12 M dithiothreitol,
12% sucrose, 4% sodium dodecyl sulfate (SDS) and 0.06%
bromophenol blue, denatured at 100˚C for 3 min and loaded on
15% SDS-PAGE.
Results
Presence of Hydrogen Peroxide on the Synechococcus elongates Strains ECT and ECT16-1
In order to test whether sHSP plays a role in the oxidative
stress response, we compared the effect of hydrogen peroxide
on the ECT16-1 strain harboring a Synechococcus vulcanus
hspA expression vector with that on the ECT strain, the reference
strain, which harbors the vector without the hspA gene.
Chlorophyll curves for ECT and ECT16-1 in the presence of 1
mM hydrogen peroxide at 30˚C under a light intensity of 30 μE
m-2/s-1 are shown in (Figure 1) revealed that the ECT strain
had much lower chlorophyll than the ECT16-1 strain. These results
suggest that HspA enhances cell survival and tolerance to
peroxide stress.
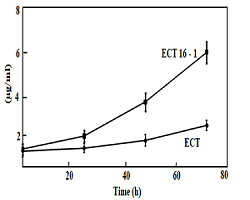
Figure 1: Cell growth of the Synechococcus ECT and and ECT16-1 strains incubated in the presence of 1mM hydrogen peroxide for 0, 24, 48 and 72 h. Cells were cultured at 30°C absorbance at 730 nm studied at 0 h. Hydrogen peroxide was added to final concentration of 1 mM after that and culture maintained up to 3 days, absorbance reading has taken every 24 h interval. Results are average of three independent experiments; bars indicate standard deviation.
Samples containing equal amount of chlorophyll were loaded
to the sucrose density gradients and the PSI complexes were
isolated and separated from ECT and ECT16-1 strains by centrifugation.
Three chromophorecontaining bands were observed,
which is consistent with a previous report. That report
showed that the upper yellow, the middle green, and the bottom
green bands contain carotenoids, PSII/PSI monomers, and
PSI trimers, respectively (Figure 2a). The upper yellow band
contained, besides others, the abundant red and orange carotenoid
protein. The middle and the bottom bands contained
PSI since the PSI subunits PsaA, PsaB, PsaD and PsaF were detected
in a CBB-stained gel after SDS-PAGE (not shown). In the
bottom band, only PsaAB proteins were detected. They were
totally absent in the 72-h hydrogen peroxide treated ECT strain
(not shown). The PSI trimer band (the bottom band) became
less intense after the 48-h incubation in ECT, indicating that it is
sensitive to oxidative stress. All three chromophore-containing
bands were greatly decreased after the 72-h incubation in ECT,
whereas ECT16-1 retained all three bands at a constant level
during the oxidative stress. Photosystem I protein also analyzed
by two dimension SDS-PAGE from hydrogen treated ECT16-1
and ECT cells. In first and second dimension results showed,
the intensity of Photosystem protein very much reduced on 72
hours treated control ECT cells not in the hspA over expression
ECT16-1 cells (Figure 2b). In the photosystem protein such as
PsaD and PsaC were detected by second dimension and obobserved
lesser intensity (not shown).
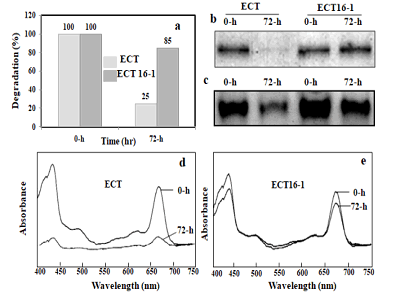
Figure 2: Photosystems protein (PsaAB) isolated by sucrose density gradient centrifugation and BN-PAGE. Cells are harvested from ECT and ECT16-1 strain incubated in the presence of 1 mM hydrogen peroxide for 0 and 72 h, respectively. Thylakoid membrane isolated and equal amount was loaded in each tube for sucrose density gradient centrifugation (250 µg of chlorophyll) and for BN-PAGE (30 µg of chlorophyll). Photosystem protein separated and analyzed by spectrophotometer (a), SDS-PAGE (b) BN-PAGE (c) double wavelength and double beam spectrophotometer for ECT (d), and ECT16-1 (e).
Characterization of Photosystem Protein
The photosystem protein (PsaAB) characters studied in the
presence of different concentration of hydrogen peroxide (0.03
to 0.3%) incubated at 4 to 37ºC for 9 h and the optimum density
was measured at 663 nm by the spectrophotometer. The
result showed three different changes, no change on lower
concentration hydrogen peroxide and low temperature. Higher
changes were observed on higher concentration of hydrogen
peroxide and higher temperature. The optimum density values
of the photosystem sample not chanced in the absence of hydrogen
peroxide at 4 and 37ºC and 0.03% of hydrogen peroxide
at 37ºC. The optimum density values of the samples reduced
20% on 0.3% hydrogen peroxide at 4ºC incubated sample and
0.1% hydrogen peroxide at 37ºC for 9 h incubation samples.
There was 65% of optimum density values was reduced when
the sample mixed with 0.3% hydrogen peroxide at 37ºC for 9 h
incubation (Figure 3a).
Figure 3: Degradation of PSI in the presence of hydrogen peroxide. PSI proteins were mixed with different concentrations of hydrogen peroxide, incubated at 0 or 37ºC for 9 h. After incubation the samples were centrifuged and supernatant was analyzed for light absorbance. Number 1 for three samples, first sample incubated at 0ºC temperature without addition of hydrogen peroxide, second sample incubated at 37ºC without addition of hydrogen peroxide and third sample incubated at 37ºC temperature with 9.6 mM of hydrogen peroxide. Number 2 for sample incubated at 0ºC temperature with 96 mM of hydrogen peroxide. Number 3 for sample incubated at 37ºC temperature with 32 mM of hydrogen peroxide. Number 4 sample incubated at 37ºC temperature with 96 mM of hydrogen peroxide (a). PSI proteins were mixed with different concentrations of hydrogen peroxide, incubated at 30ºC for 3 h. Then the samples were centrifuged and supernatant (5 μg of protein) was loaded in each well of 15% SDS/ PAGE with 7 M urea. Lance 1 for marker, 2 for a control without hydrogen peroxide treatment, lanes 3-8 for sample mixed with 16, 32, 48, 64, 80, 96, 112, 128 mM of hydrogen peroxide (b). PSI proteins were mixed with 96 mM of hydrogen peroxide, and incubated at 30ºC for 3 h. Then centrifuged sample (5μg of protein) was loaded in each well of 15% SDS/PAGE with 7 M urea and the PsaB protein was detected by Western blot using antibody. Lane 1 for control, without addition of hydrogen peroxide. Lane 2 for sample with hydrogen peroxide (c).
The photosystem protein PsaAB treated with 0.05 to 0.4% hydrogen
peroxide and incubated at 30ºC for 3 h to study the
protein degradation and aggregation. Here our results showed
the protein degradation correlated with concentration hydrogen
peroxide. We found two new degradation product bands
at 40 and 13 kDa size respectively, and the intensity of those
bands increased in higher concentration hydrogen peroxide
(Figure 3b). The degraded photosystem protein by hydrogen
peroxide was further detected by western blot analysis using
antibody (Figure 3c).
Chaperone Activity between PsaAB and HSP
The photosystem protein mixed with lysozyme for control,
α-crystallin (homologus of small heat shock protein) and small
heat shock protein HspA in one:one ratio, incubated 30ºC for
3 h in the presence of 0.3% of hydrogen peroxide to study the
chaperone activity. Here our results showed control photosystem
protein without addition of either hydrogen peroxide nor
small shock protein was affected even in the presence very low
concentration of hydrogen peroxide 0.05%. In the presence of
lysozyme control protein the photosystem protein tolerated
upto 0.15% hydrogen peroxide, after that it was degraded. By
α-Crystallin approximately 50% of photosystem protein was
protected even in the presence of high concentration of hydrogen
peroxide (0.4%). Around 80% of the photosystem protein
protected by HspA at 0.4% hydrogen peroxide (Figure 4a). One
of the major photosystem protein (PsaB), degradation was prevented
by α-crystallin in the presence of 0.3% hydrogen peroxide
at 30ºC for 3 h proved further by western blot analysis
(Figure 4b).
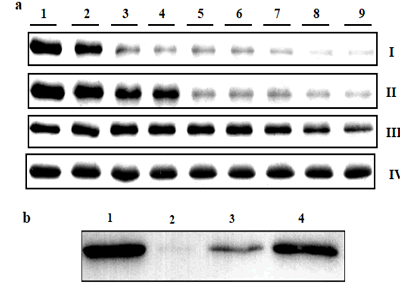
Figure 4: Direct interaction of PsaAB with HSP in the presence of hydrogen peroxide studied by SDS-PAGE (a) and Western Blott (b). PSI was mixed with different concentrations of hydrogen peroxide. Lane 1 for control without addition of hydrogen peroxide, lanes 2-9 addition of hydrogen peroxide 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35, 0.4%. First row (I) for control (photosystem sample mixed with hydrogen peroxide), Second row (II) for samples mixed with a control protein (Lysozyme), third row (III) for sample mixed with α-crystallin and fourth row (IV)for sample mixed with HspA. Sample mixed with lysozyme, or α-crystallin or HSP in one:one ratio, and incubated at 30ºC for 3 h. The centrifuged samples (5 μg of protein) were loaded on 15% SDS/PAGE with 7 M urea. Photosystem sample mixed with BSA or α-crystallin in one:one ratio, incubated at 30ºC for 3 h, centrifuged sample (5µg of protein) were loaded on 15% SDS-PAGE with 7M urea then photosystem protein (PsaB) detected by western blott analysis. Lane 1 for control sample without addition of hydrogen peroxide, lane 2 for sample with hydrogen peroxide, lane 3 for sample mixed with BSA and lance 4 for sample mixed with α-cyrstallin (b).
The above results showed that PSI proteins were protected by
α-crystallin and HspA. These results were further confirmed by
light absorbance studies. The PSI proteins were mixed with lysozyme
or α-crystallin or HspA in one:one ratio, incubated at
37ºC for 2 h in the presence of 96 mM hydrogen peroxide to
study the interaction of PSI and HSPs by a double wavelength
and double beam spectrophotometer. In the case of the PSI
proteins mixed with HspA (Figure 5a) or α-crystallin (Figure 5b),
The Chlrophyll absorption at 680 nm was kept at the same level
as the control sample (without addition of hydrogen peroxide
or α-crystallin or HspA, incubated at 0ºC). The control protein
lysozyme did not protect just like no addition sample (without
addition of α-crystallin or HspA, only hydrogen peroxide).
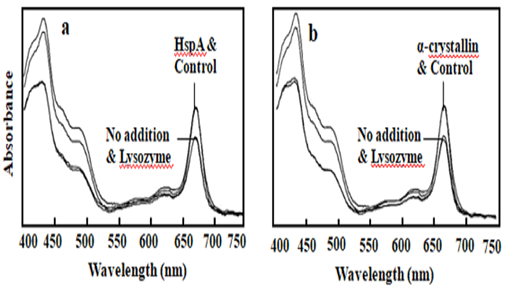
Figure 5: Light absorbance studies were undertaken for interaction of PSI and HPS in the presence of hydrogen peroxide. PSI was mixed with one:one ratio of lysozyme or a HSP, such as HspA (a) and α-crystallin (b) in the presence of 96 mM of hydrogen peroxide, incubated at 37ºC for 2 h. Then the samples were centrifuged and the supernatant analyzed for light absorbance. Control sample without addition of hydrogen peroxide and no addition sample contained only hydrogen peroxide. In the figure (a) number 1 for sample mixed with HspA and control without addition of hydrogen peroxide, number 2 for addition of hydrogen peroxide, but without addition of any protein and addition of control protein lysozyme. On the figure (b) number 1 indicates for sample mixed with α-cystallin and control sample without addition of hydrogen peroxide, number 2 indicates for addition of hydrogen peroxide, but without addition of protein and sample mixed with control protein lysozyme.
Discussion
Several heat shock proteins were induced by hydrogen peroxide.
Heat stress stimulates hydrogen peroxide generation
in plants [19]. Moreover, heat shock proteins are involved in
enhancing survival following oxidative stress in yeast, animals,
and plants [20-22]. Thus, the induction of genes encoding heat
shock proteins and a heat shock transcription factor by hydrogen
peroxide may lead to increased tolerance of further oxidative
stress, as in tomato (Lycopersiconesculentum) cells [20], as
well as contributing to tolerance of other stresses such as high
temperature [19]. It was reported that treatment with hydrogen peroxide was more effective in inducing the synthesis of
sHSPs that treatment with MV [20]. Hydrogen peroxide is able
to diffuse freely through membrane [23].
Oxidative stress causes the proliferation of peroxisomes [24].
A dense population of peroxisomes might be highly efficient in
scavenging of Reactive Oxygen Species (ROS), especially hydrogen
peroxide, which diffuses into peroxisomes from the cytosol.
Under normal growth conditions, the production of ROS in
cells is low (240 μM S-1O2- and a steady-state level of 0.5 μM
hydrogen peroxide in chloroplasts), many stresses that disrupt
the cellular homeostasis of cells enhance the production of
ROIs (240-720 μM S-1 O2- and a steady-state level of 5-15 μM
hydrogen peroxide) [25].
Photosystem Protein (PsaAB)
Photosystem proteins are very much damaged by oxidative
stress condition. Small Hsp prevents irreversible aggregation
reactions and keeps protein on the protective folding pathway.
During stress, the photosynthetic apparatus might be
suppressed or damage. In the present study when we mixed
the photosystem sample with α-crystallin the aggregation was
prevented after 48 hours dialysis, but not in the sample mixed
with Bovin Serum Albumin. The PsaA protein had a significantly
higher degradation rate than its partner in the PSI reaction center
heterodimer, PsaB. So far, it has been shown that in higher
plants and algae, the stability of the PsaB protein is more critical
than that of PsaA. For example, in Chlamydomonas reinhardtii,
it has been demonstrated that in the absence of the synthesis
of the PsaB protein, PsaA cannot be detected whereas in the
absence of the synthesis of PsaA, one can still detect the PsaB
protein in the thylakoid membrane [26].
Conclusion
Sonoike demonstrated specific degradation of the PsaB protein
during photoinhibition of PSI in spinach thylakoid membranes.
In the present study the PsaB intensity was very much
reduced by lyzosome control protein mixed sample, but not
by the HspA mixed sample. Our data demonstrate that under
oxidative stress condition induces partial damage of the PSI
complex in the Cyanobacterium Synechococcus sp. PCC 7942
control strain.
Acknowledgements
I thank Prof. Hitoshi Nakamoto, Department of Biochemistry
and Molecular Biology, Saitama University for doing this research
in his laboratory. I am also grateful to Department of
Science and Technology (DST) under Promotion of University
Research and Scientific Excellence (PURSE) Phase II program
has given me a position as Research Associate during the manuscript
writing.
REFERENCES
- Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiology Plant Mol Biol. 42: 579-620.
[Cross Ref] [Google Scholar]
- Caslake LF, Gruber TM, Bryant DA (1997) Expression of two alternative sigma factors of Synechococcus sp. Strain PCC 7002 is modulated by carbon and nitrogen stress. Microbiol. 143:3807-3818.
[Cross Ref] [Google Scholar] [Pubmed]
- Kanesaki Y, Suzuki I, Allakhverdiev SI (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp PCC 6803. Biochem Biophy Res Commun. 290: 339-348.
[Cross Ref] [Google Scholar] [Pubmed]
- Asadulghani, Nitta K, KanekoY (2004) Comparative analysis of the hspA mutant and wild-type Synechocystis sp. strain PCC 6803 under salt stress: Evaluation of the role of hspA in salt-stress management. Arch Microbiol. 182:487-497.
[Cross Ref] [Google Scholar] [Pubmed]
- Chitnis PR, Nelson N (1991) Molecular cloning of the genes encoding two chaperone proteins of the Cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem.266:58-65.
[Cross Ref] [Google Scholar] [Pubmed]
- Hossain MM, Nakamoto H (2003) Role of the cyanobacterial HtpG in protection from oxidative stress. Curr Microbiol. 46:70-76.
[Cross Ref] [Google Scholar] [Pubmed]
- Halliwell B, Gutteridge JM (1990) The antioxidants of human extracellular fluids. Arch Biochem Biophys. 280:1-8.
[Cross Ref] [Google Scholar] [Pubmed]
- Miller RA, Britigan BE (1997) Role of oxidants in microbial pathophysiology. Clin Microbiol Rev. 10:1-8.
[Cross Ref] [Google Scholar] [Pubmed]
- Smith TC (2000) Membrane heredity and early chloroplast evolution. Trends Plant Sci. 5:174-82.
[Cross Ref] [Google Scholar] [Pubmed]
- Fromme P, Mathis P (2004) Unraveling the photosystem I reaction center: a history, or the sum of many efforts. Photosynth Res. 80:109-24.
[Cross Ref] [Google Scholar] [Pubmed]
- Grotjohann I, Fromme P (2005) Structure of cyanobacterial photosystem I. Photosynth Res. 85:51-72.
[Cross Ref] [Google Scholar] [Pubmed]
- Heckathorn SA, Downs CA, Sharkey TD (1998) The small methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 116:439-444.
[Cross Ref] [Google Scholar] [Pubmed]
- Li H, Singh AK, McIntyre LM (2004) Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp strain PCC 6803. J Bacteriol 186:3331-3345.
[Cross Ref] [Google Scholar] [Pubmed]
- Herranen M, Tyystjärvi T, Aro EM (2005) Regulation of photosystem I reaction centre genes in Synechocystis sp. Strain PCC 6803 during Light acclimation. Plant Cell Physiol 46:1484-1493.
[Cross Ref] [Google Scholar] [Pubmed]
- Hihara Y, Kamei A, Kanehisa M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13:793-806.
[Cross Ref] [Google Scholar] [Pubmed]
- Muramatsu M, Hihara Y (2003) Transcriptional regulation of genes encoding subunits of photosystem I during acclimation to high-light conditions in Synechocystis sp PCC6803. Planta. 216:446-453.
[Cross Ref] [Google Scholar] [Pubmed]
- Tu CJ, Shrager J, Burnap RL (2004) Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. Strain PCC 6803. J Bacteriol. 186:3889-3902.
[Cross Ref] [Google Scholar] [Pubmed]
- Sonoike K (1996) Degradation of psaB gene product, the reaction center subunit of photosystem I, is caused during photoinhibition of photosystem I, is caused during photoinhibition of photosystem I: possible involvement of active oxygen species. Plant Sci. 115:157-164.
[Cross Ref] [Google Scholar]
- Dat JF, Lopez-Delgado H, Foyer CH (1998) Parallel changes in H2O2 and caralase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedling. Plant Physiol. 116:1351-1357.
[Cross Ref] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y (1998) Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J. 13:519-527.
[Cross Ref] [Google Scholar] [Pubmed]
- Godon C, Lagniel G, Lee J (1998) The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 273:22480-22489.
[Cross Ref] [Google Scholar] [Pubmed]
- Finkel T, Holbrook NJ (2000) Oxidant, Oxidative stress and the biology of ageing. Nature. 408:239-247.
[Cross Ref] [Google Scholar] [Pubmed]
- Levin A, Tenhaken R, Dixon R (1994) H2O2 from oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 79:583-593.
[Cross Ref] [Google Scholar]
- Lopez HE, Charlton WL, Johnson B (2000) Stress induces peroxisome biogenesis genes. EMBO J. 19:6770-6777.
[Cross Ref] [Google Scholar] [Pubmed]
- Desikan R, Mackerness AHS, Hancock JT (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127:159-172.
[Cross Ref] [Google Scholar] [Pubmed]
- Stampacchia O, Girard BJ, Zanasco JL (1997) A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell 9:773-782.
[Cross Ref] [Google Scholar]
Citation: Citation Sakthivel K (2022) Photosystem Proteins Protects by Small Heat Shock Protein in Cyanobacteria under Oxidative Stress Condition. Biochem Mol Bio J. 08:55.
Copyright: Sakthivel K. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.






