Research Article - (2022) Volume 9, Issue 12
Pathogenesis of SARS-CoV-2 and its Variants
Murtaza Bohra1,
Uma Rani1 and
N Murugan2*
1Department of Microbiology, Infectious Diseases-Molecular Genetics Division, Mahindra University, Hyderabad, Telangana, India
2Department of Biotechnology, Infectious Diseases-Molecular Genetics Division, Dr. M.G.R. Medical University, Tamil Nadu, India
*Correspondence:
N Murugan, Department of Biotechnology, Infectious Diseases-Molecular Genetics Division, Dr. M.G.R. Medical University, Tamil Nadu,
India,
Email:
Received: 14-May-2022, Manuscript No. IPBJR-22-13452;
Editor assigned: 16-May-2022, Pre QC No. IPBJR-22-13452(PQ);
Reviewed: 30-May-2022, QC No. IPBJR-22-13452;
Revised: 20-Oct-2022, Manuscript No. IPBJR-22-13452(R);
Published:
27-Oct-2022, DOI: 10.21767/2394-3718.9.12.123
Abstract
COVID-19 is a major health concern globally. The virus found to have zoonotic transmission
with latency period of the virus is 2-14 days. These viruses majorly infect the type 2 pneumocystis
in the alveoli of lungs by binding to the Angiotensin Converting Enzyme 2 (ACE-2) with the help
of spike proteins. The virus divides and replicates using host organism’s replication machinery and get
released into the surrounding cells by exocytosis. In response to the virus, the host body release
inflammatory mediators which results in cytokine storm and leads to high fever and impaired
oxygenation.
Keywords
Cytokine storm; Hemagglutinin esterase; Small membrane; Angiotensin converting
enzyme; Nucleocapsid
INTRODUCTION
Coronaviruses called after the crown like shape on the outer
surface and infects both animals and human. It has been
divided into three genera or groups based on its serological
cross reactivity and the same was confirmed through genomic
analysis which was later used for nomenclature by the
international committee on taxonomy of viruses in 2009.
Coronaviruses contains the largest known positive strand RNA
genomes of sizes ranges 30-32 kb. The replicas locus encoded
within 5 end and the structural protein at the 3’ end of the
genome following an order of Hemagglutinin Esterase (HE)
(HE is only present in some beta coronaviruses) followed by
spike (S), small membrane (E), Membrane (M) and
Nucleocapsid (N) and Internal (I) protein, encoded within the
N gene. While assembly spike proteins arranged themselves in
trimmers forming polymers embedded in envelope forms a
structure of crown. Replicas gene contains two large motifs
namely ORF1a, ORF1b encompassing two third of the genome and on translation encodes for two large polyproteins pp1a
and pp1b respectively [1-5].
Materials and Methods
SARS-CoV-2
In December 2019, Wuhan city in China reported several
cases of pneumonia in hospital with the patient having the
symptoms of dry cough, fever. On January 7, Chinese
researchers identified a new viral disease as the origin of
acute infection. At the end of January 2019, the genomic
sequences analysis was perform between different classes of
coronaviruses. It shared 79.6% similarity to SARS-CoV and
50% with MERS-CoV sequences, followed by 96% genome
sequence similarity between Bat-CoV-RaTG13 and SARSCov-
02 which proved to be a potential suspect for
transmission of the disease, however, in January 22, 2020
after WHO delegate visit, it was strongly support as human transmission. Due to increase in number of cases worldwide
in March 2020, WHO declares it as a global public health
emergency? [6-8].
Structure
Like other coronavirus species, SARS-CoV-2 composed of four
main structural proteins namely "S" protein (Spike
glycoprotein), "N" protein (Nucleocapsid protein), "E" protein
(Envelope glycoprotein), and "M" protein (Membrane
glycoprotein) along with sixteen non-structural proteins
(nsp1-16). The primary function of spike protein of any virus is
to attach itself to the receptors available on the surface of cell
membranes of host organisms which varies from species to
species. SARS virus has a receptor binding domain that targets
a particular receptor called ACE-2 with a greater affinity rate.
Carbohydrates molecules present in spike protein facilitate
the conformational changes on binding with the receptor
which allows the virus to get inside by endocytosis. Spike
proteins consist of two functional subunit S1 and S2. S1 unit
contains N-Terminal Domain (NTD) and Receptor Binding
Domain (RBD). S2 unit comprised of Fusion Peptides (FP),
Heptad Repeat1 (HR1), Central Helix (CH), Connector Domain
(CD), Heptad Repeat2 (HR2), Trans Membrane domain (TM),
and Cytoplasmic Tail (CT). The function of S1 to bind with
receptor whereas S2 is to fuse the membrane between host
cells and viral to promote entry of viral nucleic acid into the
host cells. The site between both subunits is always referred
as S1/S2 cleavage site which is responsible for the activation
of proteins which helps in fusion of both host and viral
membranes through irreversible conformational changes. M
protein is present abundantly on the viral surface and with
other structural protein collectively decides the virus size and
the structure of the virus. Membrane protein and envelope
protein work mutually to form new viral particles. The capsid
protein envelope that encases the virus Genome resides
under the viral envelope. The primary function of N protein is
not well understood, but may be involved in the transcription
of the viral genome to make several new copies of the virus
(Figure 1) [9-13].
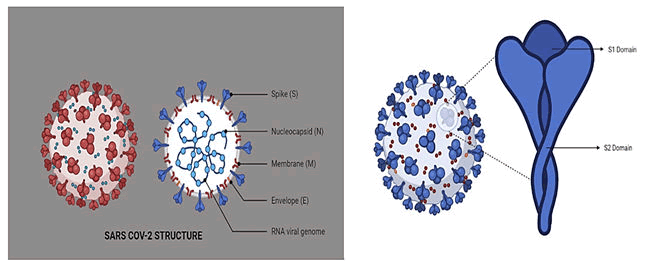
Figure 1: SARS CoV-2 structures.
Pathogenesis
The coronavirus targets and attacks the respiratory system,
which is transferred from one person to another by droplets
and fomites. The person who carries the virus either may be
symptomatic or asymptomatic. The virus has a 2–14 days
latency phase; during that time, the virus replicates itself in
the lungs. The human lungs contain sacs of alveoli which is made up of two different cells; type 1 pneumocystis and type
2 pneumocystis, type 1 primary function in alveoli is in the
exchange of gases, whereas type 2 pneumocystis help to
produce surfactant which mainly keeps alveoli open for
exchange of gases by decreasing the surface tension and
reduce the collapsing pressure. The trimmers of spike proteins
bind to ACE-2 (Angiotensin Converting Enzyme 2) which is
highly expressed on adult nasal epithelial cells and pulmonary
epithelial cells of humans. Along with S1/S2 cleavage site,
host cell surface proteins such as TMPRSS2 (Transmembrane
serine protease 2), furin (facilitates pH dependent entry) and
endosome cathepsind B and L also helps in fusion of viral
membrane into the host cell membrane. The +SSRN genome
encapsulated by nucleocapsid protein inside host cell utilizes
the host ribosomes to translate ORF region, ORF1a and ORF1b
into replicas polyprotein pp1a and pp1b respectively. Pp1a
and pp1b then undergoes proteolysis to form RTC (Replication
and Transcription Complex) and to generation number of
non-structural proteins which helps in amplification of viral
genome and also in viral assembly. RTC comprised of DMVs
(Double Membrane Vesicles), convoluted membranes and
small open double membranes which creates protection
environment for the transcription of sub genomic (+) RNA.
RTC synthesizes both (-) RNA (serves as a template for
+genomic RNA) and +RNA (sub-genomic RNA). +mRNA on
translation encodes for all the structural proteins and whole
viral assembly (both genomic DNA and structural proteins)
occurs with the help of ER-Golgi intermediate complex which
released by exocytosis to infect other cells or organisms
(Figure 2).
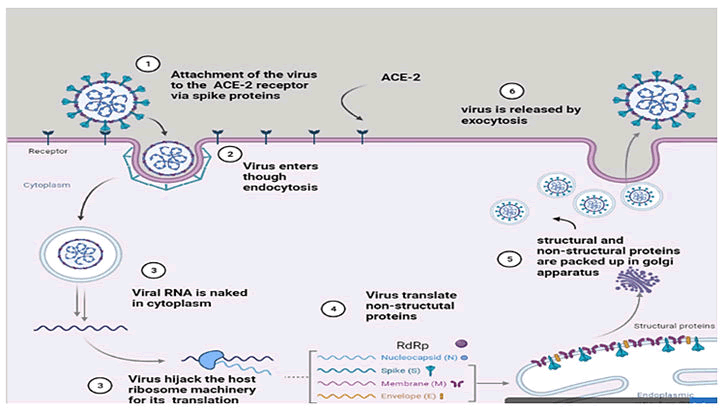
Figure 2: Illustration of pathogenesis of SARS CoV-2.
Due to the rapid replication of the virus inside the alveoli
releases inflammatory mediators, which in result alert the
macrophage, and once macrophages get stimulated, it
secretes specific cytokines to induce protection against the
virus in surrounding non infected cells. Macrophages release
cytokines such as TNF-α, interleukin-1. Apart from this,
cytokines such as interleukin-6, interleukin-8 and interleukin-1
are pro inflammatory cytokines and increase vascular
permeability and increase adhesion molecules by dilating the
endothelial cells beneath the alveolus. This leads to
vasodilation increase in capillary permeability resulting the
fluid inside leaked out into interstitial spaces and goes into
alveoli, which shrinks or compresses the alveoli. The fluid
when enters into the alveoli, drowns out all the surfactant
from the alveoli, leading to high surface tension and alveoli collapse. Due to which the patient will suffer from impaired
oxygenation known as hypoxemia. Inflammatory mediators
also attract neutrophils at the injury site and try to eliminate
the virus by releasing reactive oxygen species known as
proteases, which damages the surrounding type 1, and types 2
alveolar cells. As a result, both the gas exchange process, as
well as surfactant is decreased. IL-1 and IL-6 that are produced
in large amounts signals the hypothalamus to release
prostaglandins. Prostaglandins, IL-1, IL-6, and TNF-α, are
altogether responsible for causing fever, symptoms of
infection due to COVID [14].
Variants of SARS-CoV-2
Mutation leads to variants and types of variants are
recognizing based on the site where mutation have occurred.
Every human body and race is different from each other,
which is govern by the climate and food we food we eat and
accordingly affects our immune system. The immunological
responses of every individual varies and therefore pathogens
need to manipulate their system to be able to survive in
human body which may leads to the number of variants of
SARS-CoV-2 pathogens. Spike protein mainly consists of S1
and S2 subunits. S1 Subunits contains SP (Signal Peptide),
NTM (N- Terminal Domain), RBD (Receptor Binding Domain),
RBM (Receptor Binding Motif) whereas S2 Subunits HR1 and
HR2, TM (Transmembrane Anchor) and IC (Intracellular Tail).
As per the articles published recently, most of the mutations
have been observe in NTM and RBD of S1 subunits spike
proteins of SARS-CoV-2 pathogens, which have been
explained in details below [15].
B.1.1.7 variant
Also known as Alpha variant and was first observed in UK on
November 2020. The variant consist of total 23 mutations out
of which 8 mutations are seen in spike protein. As per the
recent studies demonstrates, three mutations plays major
role for the variation namely N501Y, spike deletion 69-70 and
P681H. N501Y mutation changes the amino acid from
Asparagine to tyrosine at position 501 in spike protein RBD
(Receptor Binding Domain) on the other hand, spike deletion
of 69-70 in the RNA genome leading to deletion of 2 amino
acid in spike proteins with unknown significance. Infect P681H
mutation changes proline to histidine at position 681 in the
spike protein which is still unclear (Figure 3).
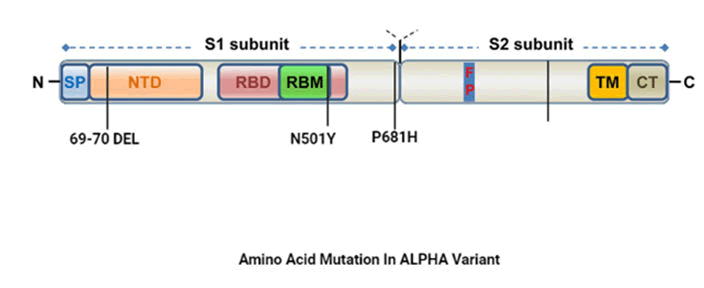
Figure 3: Amino acid mutation in alpha variant.
B.1.351 variant
It was first noticed on December 2020 in South Africa and
have labeled as a BETA variant by WHO. This variant possesses
numerous mutations with significant biological role. The
mutation is majorly observed in receptor binding region of
spike protein like N501Y in Alpha variant it forms an extra
interaction with the RBD and also increased the binding
affinity of the spike protein to ACE-2 receptor which in result
increases infectivity. E484K mutation changes glutamic acid
(E) by lysine (k) at position 484. This mutation also known as
escape mutation as it improves virus ability to evade host
immune system easily. K4179-E484K and N501Y mutation are
examples of substitution a mutation which provides an extra
but essential interaction and signifies higher infectivity in RBD
(Figure 4).
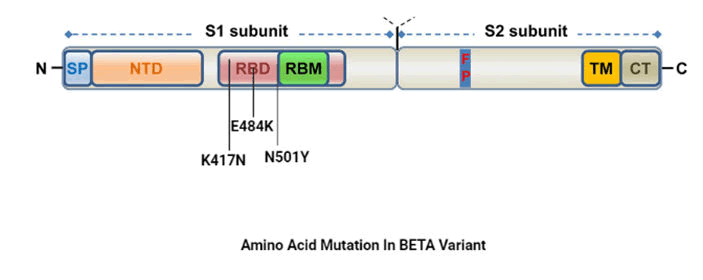
Figure 4: Amino acid mutation in beta variant.
P.1 variant
It is also known as gamma variant and was first observed in
January 2021 in Brazil. The variant comprises 17 amino acid
mutation and majorly concerned mutations are K417T, E484k
and N501Y. The variant has 8 mutations in which 4 mutations
are synonymous genetic mutation in ORF1a and ORF1b region
with one deletion mutation. Gamma has ten mutations in
spike protein including N501Y and E484K and also has two
mutations in its ORF8 region and one in N gene. Convalescent
and vaccine sera show significant loss of neutralizing activity
against P.1 lineage (Figure 5).
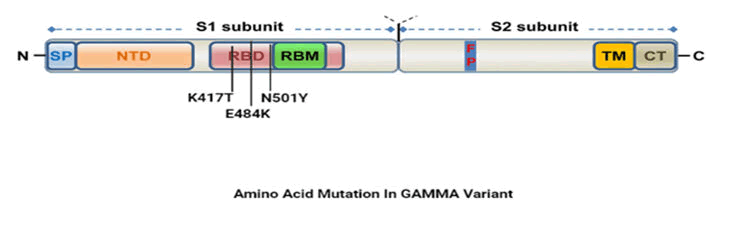
Figure 5: Amino acid mutation in gamma variant.
B.1.617.2 variant
Also known as delta variant and was first observed in India on
October 2020 namely L452R and E484Q. L452R substitute
Leucine (L) to arginine (R) at position 452 in spike protein
which modifies the virus affinity for the ACE-2 receptor and
decrease recognition capability of immune system. Another
mutation, E484Q generally substitute glutamic acid (E) to
glutamine (Q) at position 484 in spike protein resulting in
stronger binding potential to the human ACE-2 receptor as well as better ability to evade host immune system as
compare to other variant (Figure 6).
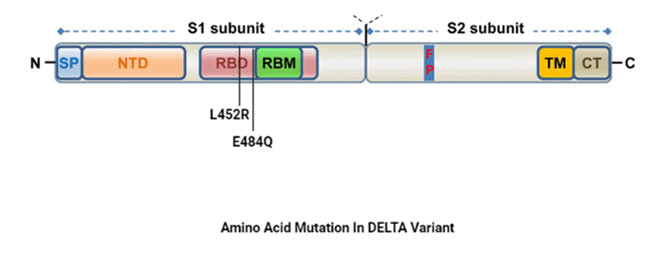
Figure 6: Amino acid mutation in delta variant.
Results and Discussion
B.1.427 variant
Also known as epsilon variant and was first discovered on
July 2020 in California USA. The variant has 5 defining
mutation (14205V and D1183Y in the ORF1ab gene and
S131, W152C, L452R in the spike protein). Epsilon is
considered to be more transmissible than previous
circulating variants. It shows about 20% increase in viral
transmissibility (Table 1).
| Variant |
Place first identified |
Date of identification |
Variant name given by WHO |
mutations |
Transmissibility |
| B.1.1.7 |
United kingdom |
Sep 2020 |
Alpha |
N501Y 69-70DEL P681H |
More than 43% compared to previous strain |
| B.1.351 B.1.351.2 B.1.351.3 |
South Africa |
May 2020 |
Beta |
N501Y K417N E484K |
More than 50% transmissible compared to previous strain |
| P.1 P.1.1 P.1.2 |
Brazil |
Nov 2020 |
Gamma |
N501Y E484K K4179 |
25-61% transmissible compare to previous strain |
| B.1.617.2 AY.1 AY.2 |
India |
Oct 2020 |
Delta |
E484Q L452R |
60% transmissible compare to previous strain |
| B.1.427/B.1.429 |
USA |
Jul 2020 |
Epsilon |
S13I W152C L452R |
20% increase in transmissibility compared to previous one |
| B.1.525 |
United Kingdom/ Nigera |
Dec 2020 |
Eta |
E484K, D614G, 69del, 70del |
Unknown significance |
| B.1.526 |
USA |
Nov 2020 |
Lota |
L452R, E484K, D614G |
Unknown significance |
| B1.617.1 |
India |
Dec 2020 |
Kappa |
L452R, E484Q, D614G |
Unknown significance |
| P.2 |
Brazil |
Apr 2020 |
Zeta |
E484Q, D614G |
Unknown significance |
Table 1: Increase in viral transmissibility.
Conclusion
The SARS-CoV-2 virus binds to the ACE-2 receptor with high
affinity as compared to the other species of coronaviruses.
Virus spike protein helps the virus to get attached to ACE2 and
get into the host cell by direct fusion or endocytosis. The virus
primarily targets type 2 pneumocystis in alveolar cells and
replicate the viral genome within the host cell. The virus is
released from the host cell by exocytosis and which infects
other surrounding cells leads to the development of a
cytokine storm in the body and cause impaired oxygenation in
the host. The immunological responses of each and every
individual are different and pathogens get adapted to the host using mutations in the genome of pathogens for survival as
well as to escape the counter attacks of host immunity.
References
- Ysrafil AI (2020) Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab Syndr. 14(4):407-412.
[Crossref][Google Scholar][Indexed]
- Sandrine B, Millet JK, Licitra BN, Whittaker GR (2012) Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses. 4(6):1011-1133.
[Crossref][Google Scholar][Indexed]
- Bian, Jingwei, Zijian Li (2021) Angiotensin Converting Enzyme 2 (ACE2): SARS-CoV-2 Receptor and RAS Modulator. Acta Pharmaceutica Sinica B. 11(1):1-12.
[Crossref][Google Scholar][Indexed]
- Ruiz C, Victor J, Montes RJ, Jose M, Puerta P, et al. (2020) SARS-CoV-2 Infection: The Role of Cytokines in COVID-19 Disease. Cytokine Growth Factor Rev. 54:62-75.
[Crossref][Google Scholar][Indexed]
- Xiaowei L, Geng M, Peng Y, Meng L, Shemin L (2020) Molecular Immune Pathogenesis and Diagnosis of COVID-19. J Pharm Anal. 10(2):102-108.
[Crossref][Google Scholar][Indexed]
- Neuman WB, Kiss G, Kunding HA, Bhella D, Baksh MF, et al. (2011) A Structural Analysis of M Protein in Coronavirus Assembly and Morphology. J Struct Biol. 174(1):11-22.
[Crossref][Google Scholar][Indexed]
- Pathoulas JT, Stoff BK, Lee KC, Farah RS (2020) Ethical Outpatient Dermatology Care during the Coronavirus (COVID-19) Pandemic. J Am Acad Dermatol. 82(5):1272-1273.
[Crossref][Google Scholar][Indexed]
- Ivan S, Agrawal R (2020) Can the Coronavirus Disease 2019 (COVID-19) Affect the Eyes? A Review of Coronaviruses and Ocular Implications in Humans and Animals. Ocul Immunol Inflamm. 28(3):391-395.
[Crossref][Google Scholar][Indexed]
- Supaporn W, Tan CW, Maneeorn P, Duengkae P, Feng Zhu (2021) Evidence for SARS-CoV-2 Related Coronaviruses Circulating in Bats and Pangolins in Southeast Asia. Nat Commun. 12(1):972.
[Crossref][Google Scholar][Indexed]
- Peng Z, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020). A Pneumonia Outbreak Associated with a New Coronavirus of Probable Bat Origin. Nature. 579(7798):270-273.
[Crossref][Google Scholar][Indexed]
- Tyrrel DAJ, Almedia JD, Berry DM, Cunningham CH, Hamre D, et al. (1968) Coronavirus. Nature 220:650.
- McIntosh K (1974) Coronaviruses: A comparative review. Curr Top Microbiol Immunol. 63:85-129.
[Google Scholar]
- Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L (2003) A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 148:2207-2235.
[Crossref][Googlescholar][Indexed]
- Sommer IE, Bakker PR (2020) What can psychiatrists learn from SARS and MERS outbreaks? Lancet Psychiatry. 7(7):565-566.
[Crossref][Googlescholar][Indexed]
- Zhou P, Yang X, Wang X (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579:270–273.
[Crossref][Googlescholar][Indexed]
Citation: Bohra M, Rani U, Murugan N (2022) Pathogenesis of SARS-CoV-2 and its Variants. Br J Res. 9:123.
Copyright: © 2022 Murugan N, et al. This is an open-access article distributed under the terms of the Creative Commons
Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and
source are credited.