Review Article - (2016) Volume 2, Issue 2
Vi T. Dang1,2 and Geoff H Werstuck1,2,3*
1Thrombosis and Atherosclerosis Research Institute, Canada
2Department of Chemistry and Chemical Biology, Canada
3Department of Medicine, McMaster University, Hamilton, Ontario, Canada
*Corresponding Author:
Geoff H. Werstuck
Thrombosis and Atherosclerosis Research Institute
237 Barton Street East, Hamilton
Ontario L8L 2X2, Canada
Tel: (905) 521-2100 ext. 40747
Fax: (905) 577-1427
E-mail: Geoff.Werstuck@taari.ca
Received Date: March 10, 2016; Accepted Date: March 20, 2016; Published Date: March 27, 2016
Citation: Dang Vi T and Geoff H. Werstuck, Metabolomics-Based Biomarkers of the Pathogenesis of Atherosclerosis Biomark J. 2016, 2:10. doi: 10.21767/2472-1646.100018
Copyright: © 2016 Geoff H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Atherosclerosis is a progressive disease of the mediumlarge arteries and the major underlying cause of most cardiovascular diseases. Despite recent advances in atherosclerosis research, the molecular mechanisms underlying the pathophysiology of this disease are not clear. Conventional methods to detect atherosclerosis are invasive, time-consuming and are associated with significant risk. The identification of metabolomics-based biomarkers of atherosclerosis would allow the development of a reliable, sensitive and non-invasive diagnostic technique. The relative levels of specific metabolites provide a direct functional readout of the physiological or pathological state of an organism and hence, can act as a biomarker of disease and/or biomarker of a specific stage of disease progression. Biomarker discovery also provides insight into established and novel metabolic pathways associated with the pathogenesis and progression of atherosclerosis. In this review, we explore recent progress in metabolomicsbased biomarker discovery in both human and animalbased atherosclerosis research. We highlight the need for direct and translational biomarkers for the diagnosis of atherosclerosis and assessment of atherosclerotic progression.
Keywords
Biomarkers; Atherosclerosis; Metabolomics; Diagnosis; Mass spectrometry; Nuclear magnetic resonance.
Abbreviations
ApoE−/−: Apolipoprotein E-deficient; CHD: Coronary heart disease; CVD: Cardiovascular disease; GC: Gas chromatograph; LC: Liquid chromatography; LDLr−/−: Low-density lipoprotein receptor deficient; MS: Mass spectrometry; NMR: Nuclear magnetic resonance.
Introduction
Cardiovascular disease (CVD) is the leading cause of mortality and morbidity worldwide [1,2]. Risk factors of CVD includes dyslipidemia, hypertension, obesity, smoking and diabetes mellitus [3]. Atherosclerosis, which is the major underlying cause of most CVDs, is a multifactorial, progressive disease of the medium-large arteries [4]. The hallmarks of this disease are the accumulation of lipid and inflammatory factors within the arterial wall that results in the hardening and narrowing of blood vessels.
Decades of research have significantly increased our understanding of the cellular mechanisms that define the pathophysiology of atherosclerosis [5–8]. Yet, despite these advances, many questions regarding the specific molecular mechanisms underlying the pathogenesis and progression of atherosclerosis remain unanswered. Furthermore, despite great advances in technologies to detect atherosclerosis, the presence of this disease is often not revealed until the latest stage, which presents as a myocardial infarction or stroke.
Current conventional methods to identify patients with atherosclerosis tend to be time-consuming and expensive [9]. Non-invasive diagnostic techniques such as carotid ultrasound and coronary artery calcium scoring only have moderate predictive values [10]. Alternately, angioplasty, which is the current gold standard of atherosclerosis detection, is invasive and carries significant risks [5,11]. As such, there is a need for a sensitive and non-invasive biomarker-based diagnostic technique.
Multiple ‘omics’-based technologies have been used to map pathways and/or mechanisms involved in disease pathogenesis in order to identify putative disease biomarkers. Genomics and proteomics are the two most established ‘omics’ platforms. These techniques have been utilized to identify several promising biomarkers of atherosclerosis including angiotensin converting enzyme [12], guanine nucleotide-binding protein β-3 [8,13], heat shock protein 27 [14,15] and suppression of tumorigenicity 2 [16,17].
Metabolomics-based technologies, the latest ‘omics’ strategy, offer a novel approach to biomarker discovery as the relative level of specific metabolites provides a direct functional readout of a physiological or pathological state of an organism [18]. Moreover, the levels of metabolites reflect the functional level more accurately than the other “omics” studies since the metabolic fluxes are regulated not only by gene expression, but also by environmental stresses [19].Therefore, the status of metabolite levels can act as the biomarkers of a disease and/or biomarkers of specific stages of disease progr20ession.
Metabolomics-based technologies are also capable of detecting early biochemical perturbation during the pathogenesis of a disease [20]. As such, predictive biomarkers can be identified to assess risks of developing the disease, or potentially facilitate the detection of the disease prior to the appearance of clinical symptoms and/or phenotypes. This allows for early identification and intervention in patients at risk, or in early stages of atherosclerotic development.
Pathogenesis and progression of atherosclerosis
Atherosclerosis is characterized by the accumulation of lipids, inflammatory factors and fibrous elements in the inner lining of the artery wall (Figure 1) [4]. The pathogenesis of atherosclerosis is initiated by the activation and/or injury at the endothelial cells, which triggers a cascade of inflammatory responses including upregulation of membrane adhesion molecules and enhancement of endothelial permeability [21]. This mediates the attachment and infiltration of blood-borne monocytes and T cells into the sub-endothelial intima. Subsequently, intimal monocytes differentiate into macrophages that endocytose cellular debris and modified lipoprotein particles giving rise to the formation of macrophage foam cells. These cellular events contribute to the development of fatty streaks, which are the hallmark features of early atherosclerotic lesions.
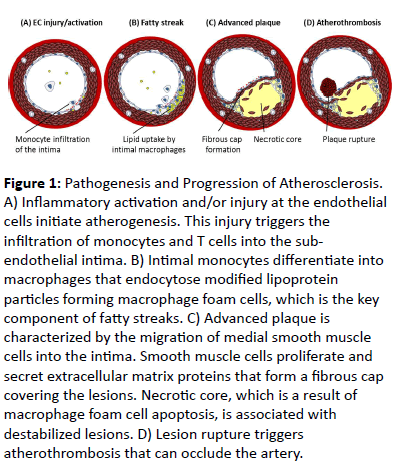
Figure 1: Pathogenesis and Progression of Atherosclerosis. A) Inflammatory activation and/or injury at the endothelial cells initiate atherogenesis. This injury triggers the infiltration of monocytes and T cells into the subendothelial intima. B) Intimal monocytes differentiate into macrophages that endocytose modified lipoprotein particles forming macrophage foam cells, which is the key component of fatty streaks. C) Advanced plaque is characterized by the migration of medial smooth muscle cells into the intima. Smooth muscle cells proliferate and secret extracellular matrix proteins that form a fibrous cap covering the lesions. Necrotic core, which is a result of macrophage foam cell apoptosis, is associated with destabilized lesions. D) Lesion rupture triggers atherothrombosis that can occlude the artery.
Within the arterial intima, lipid-engorged macrophages secrete pro-inflammatory cytokines and growth factors,intensifying the local inflammatory environment [22]. This causes the fatty streak to progress into a more advanced plaque, which is characterized by the migration of medial smooth muscle cells into the intima. Intimal smooth muscle cells then proliferate and secret extracellular matrix proteins that form a fibrous cap covering the lesions. Lipid-engorged macrophage foam cells undergo apoptosis and release internal lipid contents, leading to the formation of acellular regions called the necrotic core. Large necrotic cores are associated with destabilized lesions that are prone to rupture [23]. When lesion rupture occurs, the contents of the lesion come into contact with blood triggering atherothrombosis that can occlude the artery. Lesion rupture/atherothrombosis is the major cause of acute coronary syndrome and CV death.
Metabolomics approaches in biomarker discovery
The metabolome, which is the collection of all metabolites in a biological specimen, is an extremely complex mixture composed of metabolites from chemically diverse compound classes (e.g., amino acids, steroids, lipids, nucleosides, etc.), with different chemical properties (e.g., hydrophilic versus hydrophilic) and a dynamic range of molecular concentrations (e.g., low nM to high mM) [24]. Metabolites can undergo different types of chemical modifications including oxidation, glycosylation and methylation, which further enhance the chemical complexity in metabolomics. As such, metabolomicsbased r22esearch requires cutting-edge analytical platforms and advanced biochemoinformatic methods.
Many analytical platforms have been developed for metabolomic analyses. Nuclear magnetic resonance (NMR) spectroscopy and liquid chromatography (LC) or gas chromatograph (GC) coupled to mass spectrometry (MS) are most commonly used in metabolomics-based research [25– 27]. However, no single analytical platform is currently capable of detecting the entire metabolome. The choice of employing a specific platform is therefore dependent not only on the scope of the analysis, but also on the nature of the sample and the availability of the techniques. Ideally, multiple platforms can be used to complement each other, since they are capable of detecting different classes of metabolites.
Current advances in technology allow for the characterization of thousands of metabolites, leading to the development of so-called targeted and untargeted, or comprehensive, metabolomics (Figure 2) [28]. Targeted metabolomics involves the analysis of a specific set of known metabolites, typically focusing on one or several biological pathways of interest. This type of approach is commonly employed for hypothesis-driven research to answer specific biochemical questions, such as evaluating the effect of a drug on a specific pathway.
Figure 2: The use of metabolomics technologies in atherosclerosis research has increased rapidly. The number of Pubmed publications containing the key words “metabolomics” and “atherosclerosis” from 2005 to 2015 is shown.
Alternatively, untargeted metabolomics performs the global analysis of the metabolome and is capable of revealing novel and unanticipated molecular perturbations [28]. As such, this type of approach offers an unbiased examination of the association between the levels of metabolites and their interconnectivity in multiple metabolic pathways, in relation to a phenotype or genotype. Untargeted metabolomics is therefore commonly used as a discovery tool or a hypothesisgenerating strategy.
Depending on the scope of the analysis – targeted versus untargeted, sample preparation, analytical techniques, and data processing are susceptible to different degrees of challenge, difficulty, and scale of effort. Due to the global scope of its analysis, untargeted metabolomics produces an extremely complex data set that makes the manual examination of the metabolite features impractical. The data processing in this metabolomics approach, therefore, requires the assistance of biochemoinformatic tools.
Applications of metabolomics in atherosclerosis research
The incorporation of metabolomics-based studies in atherosclerosis research is a relatively new field of study that has expanded rapidly within the past decade (Figure 3).
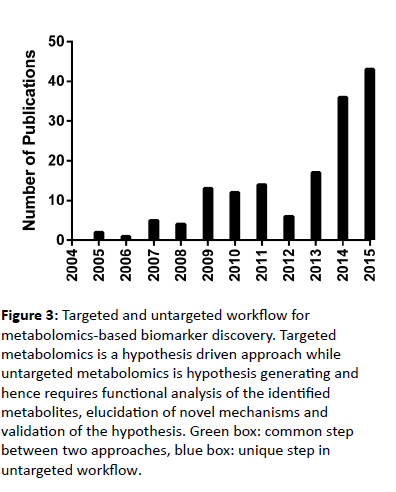
Figure 3: Targeted and untargeted workflow for metabolomics-based biomarker discovery. Targeted metabolomics is a hypothesis driven approach while untargeted metabolomics is hypothesis generating and hence requires functional analysis of the identified metabolites, elucidation of novel mechanisms and validation of the hypothesis. Green box: common step between two approaches, blue box: unique step in untargeted workflow.
Animal studies: A variety of small and large experimental animal models have been used to investigate the pathogenesis and progression of atherosclerosis. Murine models are currently the most extensively used due to the relatively low costs, ease of genetic and environmental manipulation, and relatively short time frame for atherosclerotic development [29].
In a study conducted in apolipoprotein E-deficient (ApoE−/ −) mice, choline, taurine, glycine and glucose were identified as potential biomarkers for the early diagnosis of atherosclerosis [30]. NMR analyses detected alterations in the levels of these metabolites in mice at four weeks of age, when distinct atherogenesis was evident, compared to two-week-old mice. This study also observed disturbance in alanine, methionine and valine metabolism in advanced stages of atherosclerosis. Another NMR-based metabolomics study in ApoE−/− mice suggested that urine levels of xanthine and ascorbate were potential markers of atherosclerotic plaque formation [31].
Taurocholic acid was identified as a potential plasma biomarker of early atheromatous plaque formation in a LC-MSbased study conducted in a hamster model of diet-induced atherosclerosis [32]. Taurocholic acid was up-regulated with the high-fat diet and showed strong correlation with the levels of aortic-free cholesterol. Further mechanistic studies suggested a previously unknown pathogenic role of this metabolite in early atherogenesis-associated endothelial senescence.
A comprehensive metabolomics-based study to investigate the molecular mechanisms of atherosclerosis in ApoE−/− mice was conducted by Qingbo and colleagues [33]. Both proteomic and metabolomic tools were combined to identify protein and metabolite changes in atherosclerotic vessels. NMR analysis revealed reduced levels of alanine and adenosine nucleotide in vessels of young mice (10-week-old) compared to nonatherogenic control mice. Immune activation, oxidative stress and energetic impairment were suggested to be among the earliest alterations in hyperlipidemic animals. Another commonly used mouse model of atherosclerosis is the lowdensity lipoprotein receptor deficient (LDLr−/−) mouse. Tang and colleagues employed both GC-MS and NMR-based untargeted metabolomics to analyze plasma, urine and tissues of LDLr−/− mice fed a standard or Western-type (high fat) diet [34]. Gut microbiota function, fatty acids and vitamin-B3 metabolism were found to play important roles in atherosclerotic development. Although these studies did not elucidate specific biomarkers of atherosclerosis, they did identify novel molecular pathophysiological mechanisms associated with atherosclerosis. These findings provide insight for future targeted metabolomics-based research to discover novel biomarkers associated with these identified pathways and/or mechanisms.
Human studies: Biomarker discovery in human is extremely complicated due to the genetic diversity in human population and multiple confounding factors including age, diet, medications and comorbidities. The discovery of biomarkers in human often requires the ability to differentiate subtle clinical symptoms or phenotypes, not only between patient versus control, but also within the patient group. Current diagnosis of atherosclerosis is not suitable for population-based studies because it is invasive, insensitive to early stage disease, timeconsuming and expensive. As such, biomarker discovery in this area either identified specific atherosclerosis biomarkers in small but well-defined phenotype cohorts or proposed indirect biomarkers of atherosclerosis in large epidemiological cohorts.
In a study to discover specific biomarkers of atherosclerosis, Lv and colleagues examined plasma obtained from 16 clinically evident stable atherosclerosis patients and 28 healthy controls [35]. GC-MS analyses identified palmitate, stearate and 1- monolinoleoylglycerol as potential phenotypic biomarkers for the diagnosis of atherosclerosis. This study also identified a role for palmitate in apoptosis and inflammation pathways in the development of atherosclerosis. Another small study conducted by Yu and colleagues used metabolomics-based technology to discriminate 45 patients with unstable angina from 43 atherosclerosis patients, diagnosed by coronary angiography [36]. LC-MS analyses indicated that the plasma concentrations of 16 metabolites from monoacylglycerol, tryptophan and phospholipid metabolism were altered. These metabolites showed a satisfactory diagnostic accuracy for discrimination between two groups of patients.
NMR-based metabolomics was used in the diagnosis of the presence and severity of coronary heart disease (CHD) in a study conducted by Grainger and colleagues [37]. Multivariate analyses of serum/plasma NMR spectra differentiated 36 patients with angiographically-diagnosed advanced CHD from 30 patients with normal coronary arteries, with a specificity of >90%. The predictive power of this technique was based primarily on the lipid regions of NMR spectra in a small number of patients. The same research group later stated that this technique is a weak predictor of CHD because the confounding variables of gender and drug treatments could potentially affect the lipid compositions [38]. These conflicting results further highlight the complexities introduced by variables such as diet, medications and comorbidities in human-based studies, especially in small study cohorts.
Due to the challenges in clinical diagnosis of atherosclerosis, epidemiological studies are usually employed to identify indirect biomarkers of atherosclerosis in the form of CVD risk and/or CVD mortality. In a recent Atherosclerosis Risk in Communities study, Boerwinkle and colleagues examined the longitudinal association between serum metabolome and allcause mortality among African Americans [39]. GC-MS and LCMS were used to characterize sera from 1,977 subjects with a median of 22.5 years of follow-up. CVD mortality was defined as any death in which the principle cause was cardiovascular in nature. Mannose and glycocholate were found to have strong correlation with CV mortality. The finding in this study supports the potential use of novel biomarkers beyond those associated with traditional risk factors in accessing CV risk.
A prospective study of three independent population-based cohorts employed NMR-based metabolomics to identify biomarkers for incident CVD during long-term follow-up [40]. Untargeted metabolomics was first performed on a discovery cohort with 7,256 subjects. The obtained results were then validated using targeted metabolomics in two independent cohorts with more than 2500 subjects each. The main endpoint considered in this study was the first incidence of a major CV event including myocardial infarction, ischemic stroke, cardiac revascularization or unstable angina. Phenylalanine, mono- and poly-unsaturated fatty acids were identified as biomarkers for CV risk, after correcting for confounding factors such as age, gender and other CV risk factors. The finding in this study supports the value of metabolomics-based biomarkers in improving risk assessment for CVDs.
Untargeted metabolomics was also employed to identify plasma biomarkers that predict increased risk for CVD in a translational study conducted by Stanley Hazen and colleagues [41]. Plasma from human patients who experienced a myocardial infarction, stroke or death in the ensuing three years was compared to age and gender-matched controls. LCMS analyses identified choline, trimethylamine N-oxide and betaine to be significantly associated with CVD risk. These metabolites were further validated in an independent cohort and showed significant correlations between one another, suggesting their potential interconnectivity in a common metabolic pathway. Complementary functional studies showed that they are metabolites of dietary phosphatidylcholine and implied the role of intestinal microbiome in atherogenesis. In the follow-up mechanistic studies, mice supplemented with dietary choline showed accelerated development of atherosclerosis and suppression of intestinal microflora inhibits this accelerated development. This study demonstrates that metabolomic-based technologies can be used beyond biomarker discovery and toward novel pathway/ mechanism elucidation.
Conclusion
Metabolomics-based biomarker discovery has shown potential in atherosclerosis research including assessing disease risk, diagnosing the disease, defining pathological mechanisms and identifying therapeutic targets. Emerging analytical and biochemoinformatic techniques allow a more accurate and comprehensive profiling of the metabolome. As such, multiple direct and indirect atherosclerotic biomarkers have been identified in animal models and human patients.
Animal-based studies offer more insights into the pathophysiological mechanisms of the disease and provide an opportunity to discover early disease biomarker prior to the appearance of disease phenotypes. However, due to the fundamental differences between humans and experimental animals, biomarkers discovered in animal models will have to be validated in human subjects. Alternatively, atherosclerotic biomarkers can be discovered in either small and well-defined phenotype or larger epidemiological cohorts in human-based studies. Confounding factors such as age, diet, medication and other CV risks contribute greatly to the complexity in these studies. Replication and validation of the identified biomarkers are therefore required. Perhaps ideally, human and animalbased studies can be integrated to translate the biomarker associated with the disease to the molecular mechanisms underlying that association.
Current diagnostic techniques of atherosclerosis are not only invasive and time-consuming, but also have limited value in predicting future cardiac events. Therefore, the identification of blood biomarkers of atherosclerosis will allow the development of a fast, simple and non-invasive diagnostic method. Beyond biomarker discovery, metabolomics-based technologies also provide insight into the molecular mechanisms that underlie the pathogenesis and progression of atherosclerosis. This will greatly assist in the development of effective therapeutic and preventative strategies for atherosclerosis and CVDs in general.
Acknowledgement
This research was supported by operating grants from the Canadian Institutes for Health Research (MOP62910 and MOP133612) and the Canadian Diabetes Association (OG-3-12-3852-GW).