Research Article - (2022) Volume 8, Issue 11
Genotyping of S and C Mutated Beta Globin Gene: Development of a Set of Primers for Use with Different PCR Systems
Koui Tossea A Stephane1,
Ernest Sery Gonedele-Bi2,
Eric Gbessi Adji2,
Albert A Gnondjui1,
Berenger Ako Ako1,
Coulibaly Baba1,
Offianan A Toure1,
Ibrahima Sanogo3 and
Ronan Jambou4*
1Department of Parasitology, Institute Pasteur Cote d'Ivoire, Paris, France
2Department of Parasitology, University Felix Houphouet-Boigny, Abidjan, Paris, France
3Department of Hematology, CHU Yopougon, Abidjan, Ivory Coast, Paris, France
4Department of Parasitology, Insect Vectors Institute Pasteur Paris, Paris, France
*Correspondence:
Ronan Jambou, Department of Parasitology, Insect Vectors Institute Pasteur Paris, Paris,
France,
Email:
Received: 12-Mar-2022, Manuscript No. IPBMBJ-22-12662;
Editor assigned: 15-Mar-2022, Pre QC No. IPBMBJ-22-12662(PQ);
Reviewed: 29-Mar-2022, QC No. IPBMBJ-22-12662;
Revised: 10-Oct-2022, Manuscript No. IPBMBJ-22-12662(R);
Published:
17-Oct-2022, DOI: 10.36648/2471-8084.8.12.108
Abstract
Sickle cell disease is a genetic disorder that affects nearly 5% of world population. In ivory coast, SCD
is a real problem of health and screening is not systematic after born. Here, we designed a set of
primers to detect abnormal hemoglobin S and C which can be used both in conventional and
quantitative PCR (by curves combinations analysed). A total of 60 blood samples including 13 AA, 23
AS, 9 SS, 12 SC and 3 CC hemoglobin type were screened using hemoglobin electrophoresis and PCR.
The universal control primer HBU/R4 was successfully amplified for all of 60 samples. In conventional
PCR, for typing of allele S sensibility and specificity of primers were respectively 86.36% and 87.50%.
For allele C, sensibility and specificity of this pair were respectively 53.33% and 91.11%. In qPCR,
specificity and sensitivity of primers were greater than 85% for allele S and C specific primers.
Keywords
Sickle cell disease; PCR; Ivory coast
Abbreviations
SCD: Sickle Cell Disease; PCR: Polymerase Chain Reaction; qPCR: Quantitative PCR
Introduction
Sickle cell disease is a genetic disorder that affects nearly 5%
of world population [1,2]. There are several hemoglobin
disorders known, but in Africa, most of the diseases emerge
from mutations of the codon 6 of exon 1 of the beta chain of
hemoglobin is involved is this disease. Mutations of this
codon are related to C and S types of hemoglobin associated
with the most severe disease when express as homozygote SS, CC or as hybrid SC. The mortality and morbidity associated
with SCD can be reduced if appropriate preventive measures
are undertaken. Electrophoresis of Hemoglobin (EH) is the
routine method for the diagnosis of SCD in most medical
centers. In developing countries, capillary electrophoresis is
becoming the gold standard, but this equipment is rarely used
in low income countries. Standard electrophoresis is difficult
to use because this method is invasive and be applicable after
06 months after birth due to large amount of fetal
hemoglobin [3,4]. Thus, diagnosis of SCD at maternity ward is not conducted for babies, which hinders health care. In ivory
coast, SCD is a real problem of health and screening is not
systematic after born. People are not interested in the first to
be their diagnostic cause of coast generally or the nondisponible
materiel in proximity hospital. The lack of
equipment, and the cost of the test when available, further
increases the risk for missing affected persons. Some rapid
antigenic test is on validation, but cannot give reliable
information when used in newborns. The routine method for
the diagnostic of SCD is Electrophoresis of Hemoglobin (EH) in
Cote d'Ivoire medicals centers. Molecular approaches are
more reliable, robust and non-invasive as they can be
conducted with all types of cells [5-9]. In regards to the
advantages of the molecular techniques, several authors have
used these methods to screen SCD. Here, we designed a set of
primers to detect normal hemoglobin A and abnormal
hemoglobin S and C which can be used both in conventional
and quantitative PCR (by curves combinations analysed).
These primers can be optimized for the development of rapid
isothermal molecular test.
Materials and Methods
Population, Sites of Study and Sampling
Patients were recruited at Yopougon University Hospital and Institute Pasteur Cote d'Ivoire (IPCI). After informed consent, hemoglobin status of the patients was determined using standard acetate electrophoresis. The electrophoresis of hemoglobin was realized with sebia electrophoresis kit according fabricant's protocols. For this, the sample was total blood collected in a tube with Ethylene Diamine tetraacetic Acid (EDTA). To be including in this study, the patents will be confirmed to sickle cell disease by the Electrophorese of hemoglobin (routine test). For molecular screening, the sample was Dried Blood Spot (DBS). Sample was prepared with 50 μL of total blood on 5 M whatman paper followed by drying for 24 hours at room temperature. The dried spot was stored in zip locked bag contained silicate gel for conservation until use.
Molecular Procedure
DNA purification: DNA purification was realized with DNA Blood and tissues extraction kit (Qiagen) according manufacturing protocols.
Primers Design
The sequence of human chromosome 11 (accession number NC_000011.10: c5227071-5225466 in NCBI) was used as reference sequence. The software used was Serial Clone 2.6.1 and Bio Edit 7.2.5. As positive control, a forward primer without mismatch named HBU (Hemoglobin Universal: CCTCAAACAGACACCATGGTGCA (T/C) CTGACTCCTGA) was used. To amplified specific allele A, S an d C, we designed specific forward primers called SSA (CCTCAAACAGACACCATGGTGCATC), HBS (CCTCAAACAGAACCATGGTGCA (T/C) CTGACTCCTAT) and HBC (CCTCAAACAGACACCATGGTGCA (T/C) CTGACTCCCAA). In order to make strongly the specificity of the primers for each specific a llele A, S a nd C, supplementary mismatches were introduced for S and C sequences primers (Table 1).
| Primer name |
Sequence |
Primer length (pb) |
Nucleotide substitution (accession number : NC_000011.10) |
Primer position (Conventional nomenclature) |
| HBU |
CCTCAAACAGACACCATGGTGCA(T/C)CTGACTCCTGA |
32 |
N/A |
261 to 292 |
| HBS |
CCTCAAACAGACACCATGGTGCA(T/C)CTGACTCCTAT |
35 |
G16A A17T |
261 to 292 |
| HBC |
CCTCAAACAGACACCATGGTGCA(T/C)CTGACTCCCAA |
35 |
T15C G16A |
261 to 295 |
| SSA |
CCTCAAACAGACACCATGGTGCATC |
25 |
N/A |
276 to 295 |
| R4 |
GGCAGAGAGAGTCAGTGCCTATCAGAAACCCAAGA |
35 |
261 to 285 |
| R2 |
GGCAGAGAGAGTCAGTGCCTATCAGAAACCCAAGAGTCTT |
40 |
459 to 493 |
Table 1: Sequences of primers designed.
The reverse primer R4 designed without mismatch was used as universal reverse with HBS and HBC; SSA primer was used with R3 reverse primer. All primers were used both to conventional and quantitative PCR amplification (polymerase chain reaction).
Conventional Amplification
Conventional amplification was performed in conventional PCR machine (applied bio system 9700). For HBS and HBC primers, amplification was conducted in 20 μl with 0.25 μM of each primer (specific primer/R4), molecular water, 5 x Master Mix Enzyme Ready to Load 12.5 μg MgCl2 (Solis Bio dyne and 5 μL of DNA template. Typing with SSA primer was performed in 20 μL with 2.5 mM of MgCl2, 0.25 μM of primers (SSA/R3), 0.5 mM of DNTPs and 1 U of polymerase. For each sample, four separated amplifications were conducted with SSA, HBS, HBC and HBU primers. The Initial denaturation step was performed at 95°C during 10 min followed by 35 cycles with 96°C denaturation during 10’s. Primers hybridization was conducted for 15’s at 55°C, 53°C, 56°C for HBU and HBS, SSA, and HBC respectively. The elongation was conducted at 72°C during 20’s; and 72°C during 7 min and 72°C during 7 min for the final elongation. PCR products were analysed on 2% agarose gel at 80 volts for 1 hour and visualized using geldoc imager (BIORAD).
Quantitative Amplification (qPCR)
Quantitative amplification (qPCR) was performed with rotor gene Q (Qiagen) in 20 μL containing 0.25 μM of each primer, molecular water, 5 x hotfirepol evergreen qPCR mix plus no rox (Solis Bio dyne and 5 μl of DNA template. The primer HBU was used as a positive amplification control. The PCR process started with an initial denaturation at 95°C during 10 min followed by 40 cycles with a denaturation at 96°C during 10’s.
Primers hybridization was conducted for 20’s at 65°C; and elongation at 72°C during 20’s. Both amplification and melting curves were analysed for each sample and each target (allele A, S and C). Quality of the amplification was controlled by migration of PCR product on 2% agarose gel.
Sanger Sequencing
To confirm electrophoresis of hemoglobin and PCR typing, one third of the samples were sequenced on the two DNA's strains using Sanger method (Genewiz Company).
The sequencing results were analyzed using bio edit 7.2.5 software and data were submitted on NCBI for comparison with reference sequence using BLAST.
Results
A total of 60 blood samples including 13 AA, 23 AS, 9 SS, 12 SC
and 3 CC hemoglobin type were screened using hemoglobin
electrophoresis and PCR. In this study, we considered only the
presence or not of an allele. In conventional PCR, the
universal control primer HBU/R4 was successfully
amplified for all of 60 samples. The fragment length is 233 pb
(Figure 1). 33/60 samples were amplified by the specific
primer SSA/R2. The sensibility and specificity of this primer
were respectively 91.67% and 20.63%. Using HBS/R4 specific
primers, 38/60 of the samples were amplified. The
sensibility and specificity of these primers were
respectively 86.36% and 87.50% (Table 2).
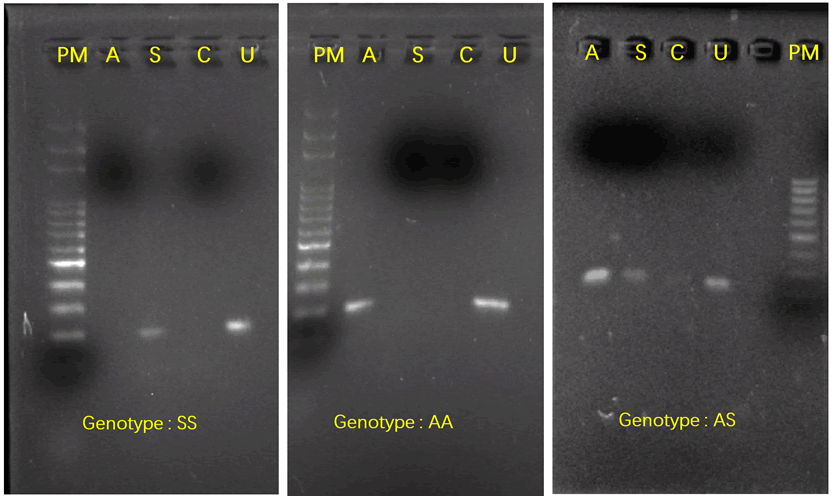
Figure 1: Amplification of the A,S and C alleles by the specific
primers SSA, HBS and HBC (respectively).
| |
Hemoglobin electrophoresis* |
Parameters |
| Sensibility |
Specificity |
| Conventional PCR (cHBB) |
| cHBB |
|
Presence of HbA |
Absence of HbA |
|
|
| Presence of allele A |
33 |
19 |
91.67%
(77.53%-98.25%) |
20.83%
(7.13%-42.15%) |
| Absence of allele A |
3 |
5 |
| |
Presence of allele C |
Absence of allele C |
|
|
| Presence of allele C |
8 |
4 |
53.33 %
(26.59%-78.73%) |
91.11 %
(78.78%- 97.52%) |
| Absence of allele C |
7 |
41 |
| |
Presence of HbS |
Absence of HbS |
|
|
| Presence of allele S |
38 |
2 |
86.36 %
(72.65%- 94.83%) |
87.50 %
(61.65%- 98.45%) |
| Absence of allele S |
6 |
14 |
| Quantitative PCR (qHBB) |
| |
Presence of HbS |
Absence of HbS |
|
|
| qHBB |
Presence of allele S |
40 |
2 |
90.91 %
(78.33%-97.47%) |
87.50 %
(61.65%- 98.45%) |
| Absence of allele S |
4 |
14 |
| |
Presence of HbC |
Absence of HbC |
|
|
| Presence of allele C |
15 |
0 |
1
(78.2%-100.0%) |
1
(92.13%-100.00%) |
| Absence of allele C |
0 |
45 |
Table 2: Amplifications parameters in this study compared to the gold standard test.
Relative to allele C typing, 8/60 samples were amplified by
specific primer HBC/R4. The sensibility and specificity of this
pair were respectively 53.33% and 91.11%. When using qPCR,
all the samples (60) were amplified by the primer HBU used
as a control (Figure 2). When amplification was conducted for
less than 21 cycles melting curves differed were 88°C. The
amplification rate was therefore 100% for the universal
primer HBU as confirmed by agarose electrophoresis of PCR
products. For amplification with primer HBS of samples with S
allele, Ct value was less than 25 cycles and hybridization
temperature was 87.5°C. 40/60 of the S containing samples
was amplified (Figure 3). In the same line, amplification and
melting curves of the primer HBC designed for abnormal
allele C, showed Ct value less than 25 cycles and hybridization
temperature at 87.3°C for the samples having at least one C
allele. Amplification was 15/60 of samples with C allele
(Figure 4). For S and C amplification, migration of PCR
products on agarose gel confirmed that only samples with Ct
less than 25 cycles harbored a true PCR band. Ct more than 25
is thus not specific. Specificity and sensitivity of primers were
greater than 85%. For the primer SSA targeting allele A, the
same problem occurred. Ct value below 20 cycles are
observed in patients with at least one A allele, but when Ct
was more than 20 cycles amplification was not specific.
Specificity of the couple SSA/R3 was than very low and needs
to be optimized. Analyse of Sanger sequences showed
correlation between electrophoresis results and phonotypical
results.
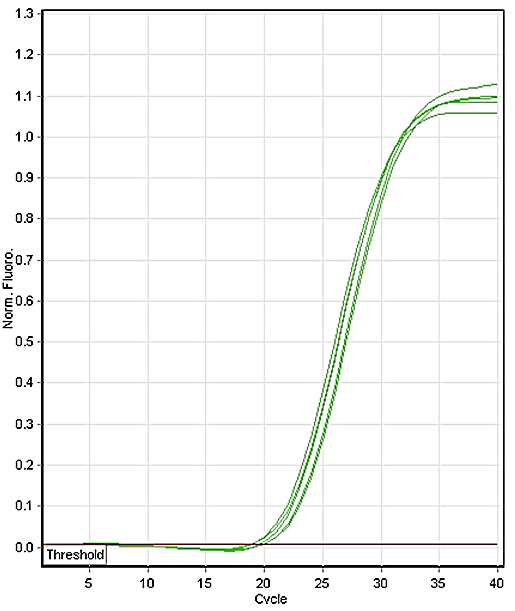
Figure 2: Amplification curve with HBU.
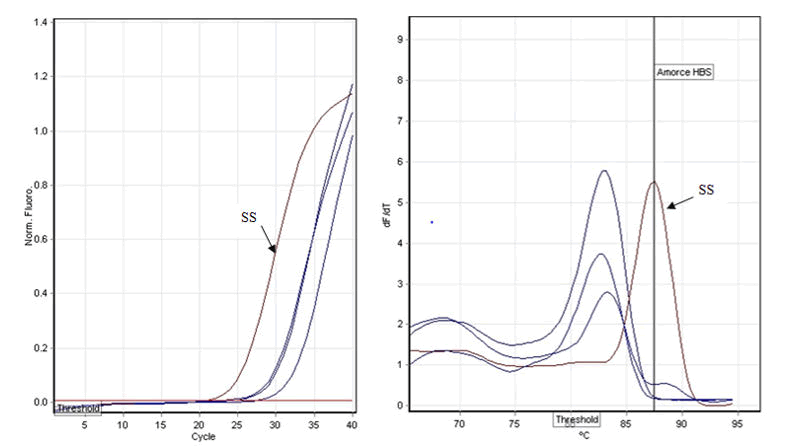
Figure 3: Amplification plot with HBS.
At left, amplification curves. Before 25 cycles, we have the
Ct of the positive sample (SS); after 25 cycles we have sample
without allele S. At right, we have melt curves. Sample with
allele S have a melt at 87.3°C and samples without allele
S have different melt temperatures.
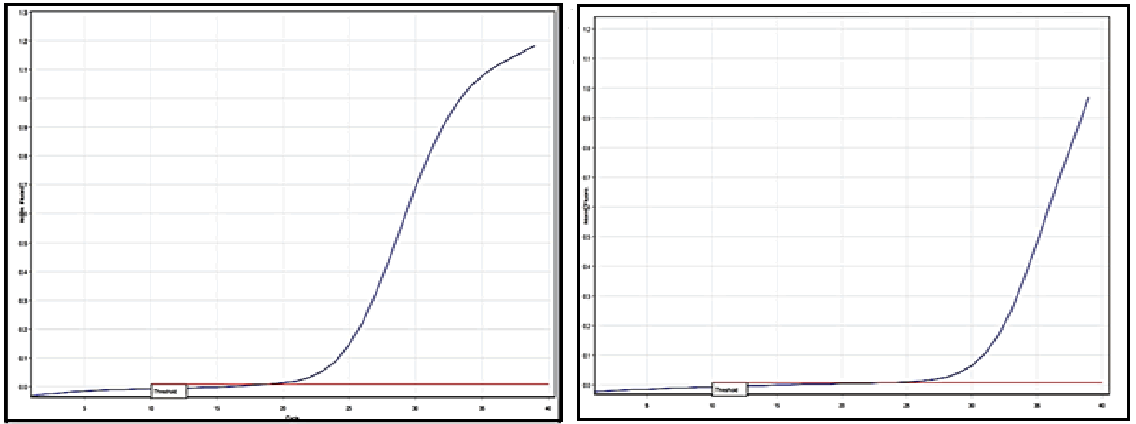
Figure 4: Amplification plot with HBC.
At left, amplification curves. Before 25 cycles, we have the
Ct of the positive sample (SS); after 25 cycles we have sample
without allele S. At right, we have melt curves. Sample with
allele S have a melt at 87.3°C and samples without allele
S have different melt temperatures At left, amplification curve of positive sample (CC). The
threshold is before 25 cycles, At right, amplification curve of
sample without allele C. The threshold is after 25 cycles.
Discussion
Primers design and detection of SNPs located on the same
codon
The aim of this study was to develop a unique molecular
primer set to be use in all PCR systems. We optimized SCD
molecular typing protocols from the designed primers. The
major difficulty was that the mutations leading to S and C
genotypes were located in the same codon. Conventional PCR
amplification can detect alleles S and C with high sensitivity
and specificity. However difficulties rose from the specificity
of primer SSA (for the typing of allele A) as it amplified all
hemoglobin allele types. That difficulty is probably caused by
a small number of mismatches between normal and abnormal
alleles which can be corrected by DNA polymerase during
amplification process. In order to increase the specificity of
primers by keeping mismatches, it could be better to use
enzymes of low fidelity which will not be able to correct these
mismatches. In the same line using master mix cannot allow
to decrease MgCl2
to increase specificity. To increase specificity of the primers,
mismatches were inserted in the sequences to decrease
stability of hybridization on the target DNA. Several authors
have tested the influence of mismatches at specific positions
on the primer on amplification and according as well as
position of the mutations in a primer sequence could
negatively influence gene amplification [10-12]. They
concluded that SNPs at the 5' end of the primer and in middle
of the sequence have very little influence on the quality of the
amplification while mutations at the 3' end negatively
affected amplification or even inhibit it. Mutations introduced
at the 3' end and another ten bases forward of the end were
the most effective. In qPCR, in order to determine the
genotypes of individuals for the S and C alleles, it was possible
to encounter both amplification curve and melting curve.
Therefore, a melting step should be included when editing the
amplification program of amplification. However, only
samples with Ct value less than 25 cycles must be considered.
Although primer designs vary between amplification methods
[13–15], we have shown here that the same set of primers
can be used for several PCR amplification methods. Results of
the two methods were consistent with each other and with
hemoglobin electrophoresis.
Phenotypic versus genotypic technics, what to prefer?
Several phenotypic methods can be used as an alternative of
electrophoresis of hemoglobin used tandem Mass
Spectrometry (MS/MS) for the typing of sickle cell disease in
newborns, whereas concluded that HPLC typing was the most
efficient method for this typing. However, mass spectrometry
remains limited by a relatively low sensitivity [16]. The HPLC
method needs technical platforms that are not usually
available in routine laboratories. Several studies have also
focused on the molecular typing of abnormal hemoglobin
used real time PCR (FRET) to detect foetal blood
contamination by the maternal blood to assign the right
genotype of the child used the same method for retrospective
typing of SCD in population with malaria in Cote d'Ivoire.
Although effective, that technique requires advanced
equipment and a qualified technician. In opposite, devices for
classical PCR are now available in most of clinical laboratory
and have used classical PCR only for alleles A and S typing.
The comparison of our conventional PCR and qPCR results
indicated no difference between the sensitivity and the
specificity of allele S typing (Chi-square test, p=0.9952); but
difference existed for the sensitivity and specificity for allele C
typing (Chi-square test, p=0.01455). For this abnormal allele
(allele C), qPCR typing present good results. These results
indicate that our primers set are suitable for molecular typing
of SCD.
Conclusion
We designed a set of primers for conventional and
quantitative amplification. These primers correctly
discriminates S and C alleles in conventional PCR and qPCR.
We recommend the use of HBS and HBC primers for the
discrimination of allele S and allele C. HBU primer can be used
as amplification control primer. The next step will be to
develop a point of care molecular test that could be perform
at room temperature and read optically. Based on these first
results, a RPA test is already in design to reach this goal.
Conflict of Interest
There are no conflicts of interest
Acknowledgements
We thank people and institutions who contributed to the
realisation of this work.
References
- WHO (2006) Sickle Cell Disease in the African Region: Current Situation and the Way Forward. World Health Organazation. 1–6.
- Tolo-Diebkile A, Koffi KG, Nanho DC, Duni S, Boidy K, et al. Drepanocytose homozygote chez l’adulte ivoirien de plus de 21 ans. Cah Sante. 20(2):63–67.
[Google Scholar]
- Forget BG, Franklin Bunn H (2013) Classification of the disorders of hemoglobin. Cold Spring Harb Perspect Med. 3(2):1-12.
[Crossref][Google Scholar][Indexed]
- Maier-redelsperger M, Bardakdjian-michau J, Guilloud-bataille M (2002) Decreased morbidity in homozygous sickle cell disease detected at birth. Hemoglobin. 26(3):211-217.
[Crossref][Google Scholar][Indexed]
- Costa C, Pissard S, Girodon E, Huot D, Goossens M (2002) A One Step Real Time PCR Assay for Rapid Prenatal Diagnosis of Sickle Cell Disease and Detection of Maternal Contamination. Mol Diagnosis. 7(1):45–48.
[Crossref][Google Scholar][Indexed]
- Akanni E, Alli OA, Mabayoje VO (2013) Molecular diagnosis of hemoglobinopathies using allele-specific polymerase chain reaction in Nigeria. Am J Biotechnol Mol Sci. 3(1):24-28.
- Verhoef TI, Hill M, Drury S, Mason S, Jenkins L, et al. (2016) Non-invasive prenatal diagnosis (NIPD) for single gene disorders : cost analysis of NIPD and invasive testing pathways. Prenat Diagn. 36(7):636–642.
[Crossref][Google Scholar][Indexed]
- Tossea SK, Adji EG, Coulibaly B, Ako BA, Coulibaly DN, et al (2018) Cross sectional study on prevalence of sickle cell alleles S and C among patients with mild malaria in Ivory Coast. BMC Res Notes. 11(1):215-220.
[Crossref][Google Scholar][Indexed]
- Toye ET, Van Marle G, Hutchins W, Abgabiaje O, Okpuzor JO (2018) Single tube allele specific PCR: a low cost technique for molecular screening of sickle cell anaemia in Nigeria. Afr Health Sci. 18(4):995-1002.
[Crossref][Google Scholar][Indexed]
- Kwok S, Kellogg DE, Mckinney N, Spasic D, Levenson C, et al. (1990) Effects of primer template mismatches on the polymerase chain reaction: Human immunodeficiency virus type 1 model studies. Nucleic Acids Res. 18(4):999–1005.
[Crossref][Google Scholar][Indexed]
- Rana KD, Stewarta G, Boissinota M, Boudreau DK (2015) Influence of sequence mismatches on the specificity of recombinase polymerase amplification technology. Mol Cell Probes. 29(2):116–121.
[Crossref][Google Scholar][Indexed]
- Daher RK, Stewart G, Boissinot M, Bergeron MG (2016) Review Recombinase Polymerase Amplification for Diagnostic Applications. Clin Chem. 62(7):947-958.
[Crossref][Google Scholar][Indexed]
- Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K (2008) Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique. perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 78(6):407–421.
[Crossref][Google Scholar][Indexed]
- Stringer OW, Andrews JM, Greetham HL, Forrest MS (2018) Twist Amp Liquid: a versatile amplification method to replace PCR. Nat Methods. 15(1):395–395.
[Google Scholar]
- Boemer F, Ketelslegers O, Minon JM, Bours V, Schoos R (2008) Newborn screening for sickle cell disease using tandem mass spectrometry. Clin Chem. 54(12):2036–2041.
[Crossref][Google Scholar][Indexed]
- Rathi S, Rathi N (2017) Reliability of Screening Tests against HPLC for the Detection of Sickle Cell Disease in Betul District A Hospital Based Study from Central India. Natl J Integr Res Med. 8(6):83–86.
[Google Scholar]
Citation: Stephane KTA, Gonedele-Bi ES, Adji EG, Gnondjui AA, Ako BA, et al. (2022) Genotyping of S and C Mutated Beta
Globin Gene: Development of a Set of Primers for use with Different PCR Systems. Biochem Mol Biol J. 8:108.
Copyright: © 2022 Jambou R, et al. This is an open-access article distributed under the terms of the Creative
Commons Attribution License, which permits unrestricted Use, distribution, and reproduction in any medium, provided
the original author and source are credited.





