Yasuo Tsunaka* and Kosuke Morikawa
Department of Gene Mechanisms, Graduate School of Biostudies, Kyoto University, Yoshida-konoemachi, Sakyoku, Kyoto, Japan
Corresponding Author:
Dr. Yasuo Tsunaka
Department of Gene Mechanisms, Graduate School of Biostudies
Kyoto University, Yoshida-konoemachi, Sakyo-ku, Kyoto 606- 8501, Japan
Tel: 81757537531
E-mail: tsunaka.yasuo.3r@kyoto-u.ac.jp
Received Date: December 18, 2016; Accepted Date: December 29, 2016; Published Date: December 31, 2016
Citation: Tsunaka Y, Morikawa K. FACT Creates a Transiently Accessible Nucleosome Structure Through Integrated Reorganization Mechanism. Biochem Mol Biol J. 2016, 2:3.DOI: 10.21767/2471-8084.100029
Keywords
Nucleosomes; Histone; Transcription
Description
Eukaryotic genomes are highly condensed in chromatin, which is dominated by arrays of the basic repeating units termed nucleosome [1,2]. This packaging limits the accessibility of DNA, thus creating a barrier that plays a major role in regulating nuclear processes such as DNA transcription, replication, and repair. The orchestrated actions of histone chaperones and ATP-dependent chromatin remodelers rearrange the barrier at nucleosome or higher order levels, thereby playing critical roles in gene regulation linked to epigenetics [3-8]. Facilitates chromatin transcription (FACT) is an essential and highly conserved histone chaperone that facilitates histone replacement, nucleosome assembly and nucleosome disassembly; thus highlighting that it can both stabilize and destabilize the barrier [5-13]. In fact, Drosophila FACT counteracts the spreading of silent chromatin through histone replacement at the boundary between heterochromatin and euchromatin [10].
FACT is a heterodimer consisting of Spt16 and SSRP1 (Pob3 in yeast) proteins, each organized by several structural domains [14]. In yeast, the functions of FACT are supported by the HMG protein, Nhp6 [15,16]. Intriguingly, the FACT subunits participate in a broad range of processes including DNA transcription, replication and repair [6,13,17-19]. In spite of this broad functional spectrum, every eukaryotic species contains only one ortholog of the FACT complex, suggesting that FACT should conduct universal actions in terms of nucleosome dynamics.
Initially, human FACT promotes the eviction of an H2A-H2B dimer from histone octamer, thereby forming hexasome, which consists of an H3-H4 tetramer and one H2A-H2B dimer (Figure 1: dimer displacement mechanism) [9]. Hexasomes are proposed to facilitate transcription, which is followed by the reassembly of octameric nucleosomes. On the other hand, yeast FACT can increase nuclease sensitivity throughout nucleosome without displacing a dimer [15]. FACT also competes with DNA for binding to H2A-H2B [20,21]. Therefore, FACT is considered to enhance unwrapping of nucleosomal DNA, by blocking its contacts with H2A–H2B. These findings suggested that FACT reorganizes nucleosomes to increase DNA accessibility, while tethering the eight histone components together [21] (Figure 1). In another word, FACT interferes with histone-DNA contacts, while preserving histone-histone interactions [22]. Whereas this nicely explains how FACT prevents histone loss, it does not appear to clearly explain how the same molecule causes further disruption of nucleosome and histone variant replacement.
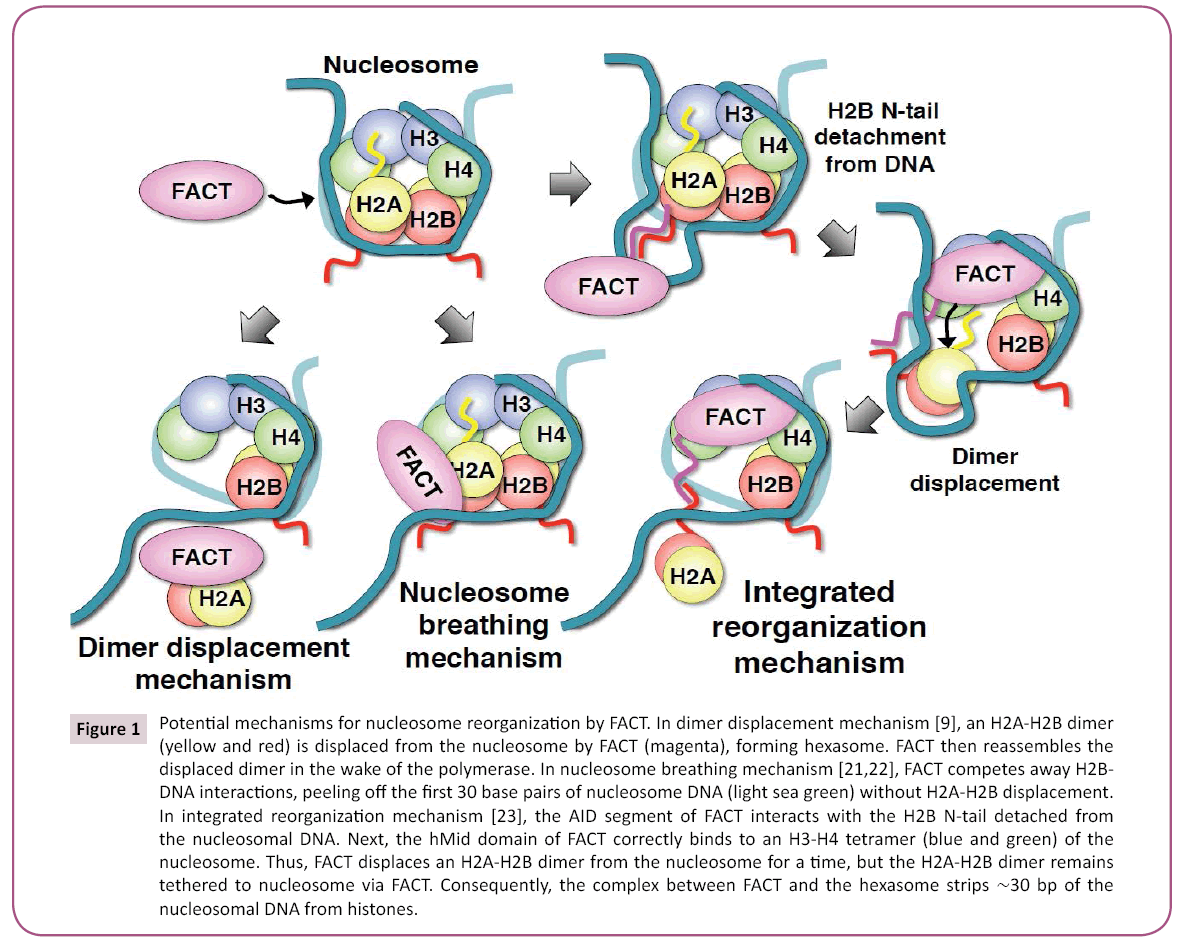
Figure 1: Potential mechanisms for nucleosome reorganization by FACT. In dimer displacement mechanism [9], an H2A-H2B dimer (yellow and red) is displaced from the nucleosome by FACT (magenta), forming hexasome. FACT then reassembles the displaced dimer in the wake of the polymerase. In nucleosome breathing mechanism [21,22], FACT competes away H2BDNA interactions, peeling off the first 30 base pairs of nucleosome DNA (light sea green) without H2A-H2B displacement. In integrated reorganization mechanism [23], the AID segment of FACT interacts with the H2B N-tail detached from the nucleosomal DNA. Next, the hMid domain of FACT correctly binds to an H3-H4 tetramer (blue and green) of the nucleosome. Thus, FACT displaces an H2A-H2B dimer from the nucleosome for a time, but the H2A-H2B dimer remains tethered to nucleosome via FACT. Consequently, the complex between FACT and the hexasome strips ~30 bp of the nucleosomal DNA from histones.
Our recent studies revealed the integrated molecular mechanism of nucleosome reorganization involving H2A-H2B displacement by human FACT [23] (Figure 1). Initially, the acidic intrinsically disordered (AID) segment of FACT interacts with a highly basic region in the H2B N-tail (HBR) detached from the nucleosomal DNA. This local disruption of DNA-histone contact may be aided by DNA torsional strain, generated by elongating polymerases [24,25], and also most likely by ATP-dependent chromatin remodelers, which create DNA loops or bulges on nucleosomes during DNA translocation [8,26,27]. Next, the rigid Mid domain of FACT correctly binds to the H2A docking surface of nucleosome. Thus, FACT displaces an H2A-H2B dimer from nucleosome for a time through steric collisions on the H2A docking surface of an H3-H4 tetramer, but the H2A-H2B dimer remains tethered to nucleosome via FACT; presumably, through transient interaction retained between the FACT AID segment and the HBR region of H2B. Consequently, the complex between FACT and hexasome strips approximately 30-bp of the nucleosomal DNA from histones. The DNA stripping may facilitate the invasion of polymerases, which must surmount the barriers against their progress [28,29]. After the relevant processes have concluded, FACT immediately reassembles the tethered dimer into nucleosome, thus remaining the eight histone components during the processes. In addition, the accessible nucleosome configuration would provide a comprehensive explanation for how FACT causes further disruption of nucleosome and edits histone variant contents through cooperation with histone chaperones, such as Asf1, HIRA, CAL1/HJURP, CAF1, Spt6, and MCM2 [10-13,30-33].
Universality of this integrated mechanism is strongly supported by some recent studies. First, Kemble et al. [34] and Valievahas et al. [16] have proposed a similar mechanism in which yeast FACT remains in the reorganized nucleosome complex and is likely to interact with both the uncoiled nucleosomal DNA and core histones, thereby replacing both of the DNA-histone and histonehistone interactions. Second, the integrated mechanism indicates that the H2B N-tail is essential for both of H2A-H2B displacement and deposition by FACT. This is verified by recent studies that the HBR region in the H2B N-tail is important for nucleosome disassembly [35] as well as assembly [36] by yeast FACT.
In conclusion, the integrated reorganization mechanism rationalizes how FACT operates to create a stable and dynamic nucleosome structure without entire nucleosome disassembly, so as to ensure chromatin integrity. However, FACT retains nucleosome in partially loosened structures by two transient interactions, in which Mid and AID of FACT bind to a H3-H4 tetramer and HBR of H2A-H2B, respectively, thereby replacing histone-histone interactions within nucleosome [23]. Therefore, it may act in concert with other factors to facilitate histone eviction and exchange.
Acknowledgment
This work was supported by Grants-in-Aid for Scientific Research (A) (JSPS KAKENHI Grant Number 23247016 and 26251008) and PRESTO, JST.
References
- Luger K, Mader A, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251-260.
- Woodcock CL, Ghosh RP (2010) Chromatin higher-order structure and dynamics. Cold Spring Harbor Perspectives in Biology 2: a000596.
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T (2013) Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490-503.
- Gurard-Levin ZA, Quivy JP, Almouzni G (2014) Histone chaperones: assisting histone traffic and nucleosome dynamics. AnnualReview of Biochemistry 83: 487-517.
- Shintomi K, Takahashi TS, Hirano T (2015) Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol 17: 1014-1023.
- Talbert PB, Henikoff S (2016) Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Biol.
- Husain A, Begum NA, Taniguchi T, Taniguchi H, Kobayashi M, et al. (2016) Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat Commun 7: 10549.
- Richard JLC, Shukla MS, Menoni H, Ouararhni K, Lone IN, et al. (2016) FACT assists base excision repair by boosting the remodeling activity of RSC. PLoS Genet 12: e1006221.
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, et al. (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090-1093.
- Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S (2007) Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev 21: 552-561.
- Heo K, Kim H, Choi S, Choi J, Kim K, et al. (2008) FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell 30: 86-97.
- Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, Robert F (2015) The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol Cell 58: 1113-1123.
- Yang J, Zhang X, Feng J, Leng H, Li S, et al. (2016) The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Reports 14: 1128-1141.
- Bondarenko MT, Maluchenko NV, Valieva ME, Gerasimova NS, Kulaeva OI, et al. (2015) Structure and function of histone chaperone FACT. MolBiol 49: 796-809.
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman DJ, et al. (2009) yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell 35: 365-376.
- Valieva ME, Armeev GA, Kudryashova KS, Gerasimova NS, Shaytan AK, et al. (2016) Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat StructMolBiol 23: 1111-1116.
- Ransom M, Dennehey BK, Tyler JK (2010) Chaperoning histones during DNA replication and repair. Cell 140: 183-195.
- Dinant C, Ampatziadis-Michailidis G, Lans H, Tresini M, Lagarou A, et al. (2013) Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol Cell 51: 469-479.
- Kwak H, Lis JT (2013) Control of transcriptional elongation. Annu Rev Genet 47: 483-508.
- Winkler DD, Muthurajan UM, Hieb AR, Luger K (2011) Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J BiolChem 286: 41883-41892.
- Hondele M, Stuwe T, Hassler M, Halbach F, Bowman A, et al. (2013) Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature 499: 111-114.
- Hsieh FK, Kulaeva OI, Patel SS, Dyer PN, Luger K, et al. (2013) Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. ProcNatlAcadSci USA 110: 7654-7659.
- Tsunaka Y, Fujiwara Y, Oyama T, Hirose S, Morikawa K (2016) Integrated molecular mechanism directing nucleosome reorganization by human FACT. Genes Dev 30: 673-686.
- Sheinin MY, Li M, Soltani M, Luger K, Wang MD (2013) Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun 4: 2579.
- Teves SS, Henikoff S (2014) Transcription-generated torsional stress destabilizes nucleosomes. Nat StructMolBiol 21: 88-94.
- Mueller-Planitz F, Klinker H, Becker PB (2013) Nucleosome sliding mechanisms: new twists in a looped history. Nat StructMolBiol 20: 1026-1032.
- Bartholomew B (2014) ISWI chromatin remodeling: One primary actor or a coordinated effort? CurrOpinStructBiol 24: 150-155.
- Chang HW, Pandey M, Kulaeva OI, Patel SS, Studitsky VM (2016) Overcoming a nucleosomal barrier to replication. Science Advances 2: e1601865-e1601865.
- Studitsky VM, Nizovtseva EV, Shaytan AK, Luse DS (2016) Nucleosomal Barrier to Transcription: Structural Determinants and Changes in Chromatin Structure. BiochemMolBiol J 2: 2.
- Takahata S, Yu Y, Stillman DJ (2009) FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34: 405-415.
- Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, et al. (2013) Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Reports 3: 892-904.
- Prendergast L, Müller S, Liu Y, Huang H, Dingli F, et al. (2016) The CENP-T/-W complex is a binding partner of the histone chaperone FACT. Genes Dev 30: 1313-1326.
- Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, et al. (2015) Establishment of centromeric chromatin by the CENP-A assembly factor CAL1 requires FACT-mediated transcription. Dev Cell 34: 73-84.
- Kemble DJ, McCullough L, Whitby FG, Formosa T, Hill CP (2015) FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide motifs. Mol Cell 60: 294-306.
- Zheng S, Crickard JB, Srikanth A, Reese JC (2014) A highly conserved region within H2B is important for FACT to act on nucleosomes. Mol Cell Biol 34: 303-314.
- Mao P, Kyriss MNM, Hodges AJ, Duan M, Morris RT, et al. (2016) A basic domain in the histone H2B N-terminal tail is important for nucleosome assembly by FACT. Nucleic Acids Research 44: 9142-9152.


