Research Article - (2025) Volume 11, Issue 5
In silico Analysis of MicroRNA of (Mentha X Piperita) Peppermint
Saqib Ishaq1*,
Jamshaid Khan2,
Raheel Tahir2,
Abdul Aziz1,
Obaid Habib3,
Muhammad Inam Ul Haq1,
Karim Gul3,
Yasir Ali4 and
Rehan Naeem2
1Department of Computer Sciences and Bioinformatics, Khushal Khan Khattak University, Methawalah, Pakistan
2Department of Biotechnology and Genetic Engineering, Kohat University of Science and Technology (KUST), Kohat, Pakistan
3Department of Biotechnology, Abdul Wali Khan University Mardan (AWKUM), Mardan, Pakistan
4Department of Biomedical Sciences, The Chinese University of Hong Kong, Ma Liu Shui, Hong Kong
*Correspondence:
Saqib Ishaq, Department of Computer Sciences and Bioinformatics, Khushal Khan Khattak University, Methawalah,
Pakistan,
Email:
Received: 19-Sep-2023, Manuscript No. IPBMBJ-23-17816;
Editor assigned: 21-Sep-2023, Pre QC No. IPBMBJ-23-17816 (PQ);
Reviewed: 04-Oct-2023, QC No. IPBMBJ-23-17816;
Revised: 07-Jan-2025, Manuscript No. IPBMBJ-23-17816 (R);
Published:
14-Jan-2025
Abstract
Mentha piperita, commonly known as peppermint, is a well-known medicinal plant with a range of therapeutic properties. In this study, we conducted an in-silico analysis of microRNAs and ESTs in Mentha piperita to gain insight into its genetic makeup. We obtained 29 EST sequences and 36 microRNAs from the NCBI database and analyzed them using various bioinformatics tools. Functional annotation of the microRNAs revealed that only 29% of the miRNAs met the criteria for AU content between 30% and 70%. Out of these, target genes were identified for only 9 sequences, which were annotated with basic roles in biological processes and molecular functions. Phylogenetic analysis confirmed the conservation of Mentha piperita with Arabidopsis thaliana. Additionally, we calculated E-values and energies for the ESTs and microRNAs, respectively. Our findings provide valuable insights into the genetic makeup of Mentha piperita and may aid in further understanding its therapeutic properties. Future research could involve the experimental validation of the predicted targets of the identified microRNAs and the characterization of the function of the ESTs.
Keywords
Mentha; EST sequence; Structural and functional annotation; Gene ontology; Phylogeny
Introduction
Mentha piperita L. (peppermint), belongs to the family Lamiaceae, one of the most significant industrial herbs with a variety of essentials oils. Mint is a member of the Lamiaceae family, which is the largest family of plants with 236 genera and 7000 species and genus Mentha, consist of 15 hybrids, cultivars, subspecies, varieties and 42 species [1]. Mentha piperita is generally called as peppermint is a perennial herb widely used by greater part of people globally in various forms like leaf, leaf extract oil and leaf water. Due to its rich flavor and fragrance peppermint has economic value both in the domestic and international market and an important export commodity that fetches good foreign revenue. The peppermint oil logs for various antispasmodic activities like antivomiting, carminativum, stomachic and antimicrobial properties. The main compounds such as menthol, menthone, and menthofuran which gives unique aroma [2]. In-silico refers to analysis which is carried out in a computer environment, rather than in the laboratory (in vitro). Plant micro RNA’s are small non-coding RNA’s that have a length of 21 to 24 nucleotides which have role in growth and in metabolic and defense progress. Mostly, miRNAs associate with the target mRNAs by 3′ UTR to curb its expression. It also interacts with gene promoters and 5′ UTR coding sequence; in addition under specific condition it activates gene expression. This miRNA can also be identified by the use of Expressed Sequence Tag (EST) and a Genome Survey Sequence (GSS). EST analysis has gained significance when compared with other approaches like conserved miRNAs and be capable of identifying without knowing the whole genome sequences. It offers straight confirmation for miRNA expression than from genomic sequence surveys without any much specified software. By this method several miRNAs are effectively recognized in diverse plant species. Many plant miRNAs are evolutionarily conserved from species to species. MiRNA precursors display high variability; their mature sequences display extensive sequence complementarity to their target mRNA sequences. MiRNAs play important roles in plant post transcriptional gene regulation by targeting mRNAs for cleavage or repressing translation [3]. MiRNAs are involved in plant development, signal transduction, protein degradation, response to environmental stress and pathogen invasion, and regulate their own biogenesis. MiRNAs regulate the expression of many important genes; a majority of these genes are transcriptional factors. Here we use homology base method meaning searching for micro RNA sequence which are involved in anti-oxidant. The specie we selected that is mentha peppermint.
Materials and Methods
Searching of EST’s in NCBI
In this study we collect total 1316 EST sequences of (Mentha X Piperita) Peppermint from National Center for Biotechnology Information NCBI. Furthermore, to screen all these 1316 EST using Mirbase online search tool with the following E-value 0.01 to 0.099 [4].
Identification of Structural and Functional Annotation
Structural annotation of these microRNA to identify their structure through online server Mfold, using Mfold to determine the best possible secondary structure and energies of these microRNA which uses energy minimization method, protein coding sequences were removed prior to prediction. Functional annotation is the main step for finding the function of obtained microRNA EST sequences [5]. Functional analysis can be done by different database here we Blast2GO database because this also give the graph along with the function of EST’s microRNA. In order to find function of the obtained EST’s sequences first you need to target the genes for the particular microRNA, because the function of microRNA is ultimately defining by the genes. For identifying the target gene for those microRNA PsRNA target software were used.
Phylogenetic Analysis
Evolutionary tree is constructed to find out relation among present sequences and their root sequences [6]. Phylogenetic tree also constructed to find out the ancestral relation among Arabidopsis thaliana and Mentha Peppermint specie. Phylogenetic tree uses a character based method called maximum likelihood that show the distance among texa. The tool that are used for the construction of phylogenetic tree is RxMI and MegaXtool, that uses the MI tree construction and its probability based approach and for construction of phylogenetic tree and evolutionary model [7].
Results
Total 1316 EST of Mentha piperita was retrieved (based on the basis of size), analyzed and annotated through the CAP3 assembly program. In the screening all the 1316 EST’s sequence only 29 miRNA sequence was retrieve [8]. After that all the 29 EST’s sequence only 36 miRNA were found meet the criteria of required E-value. We blast each EST’s sequence individually in mirbase and search for value. The result also shows and given number of A, G, C, U/T and A-T/U count are listed in Table 1.
| S. no |
Accession Number |
Sequences |
E values |
| 1 |
MIMAT0027646 |
>hsa-miR-6873-5p MIMAT0027646
CAGAGGGAAUACAGAGGGCAAU |
0.099 |
| 2 |
MIMAT0015455 |
>bmo-miR-3270 MIMAT0015455
UGUCUUUUUCCUAAAACGAUUCA |
0.036 |
| 3 |
MIMAT0009979 |
>hsa-miR-2054 MIMAT0009979
CUGUAAUAUAAAUUUAAUUUAUU |
0.042 |
| 4 |
MIMAT0030650 |
>prd-miR-7912-5p MIMAT0030650
CAUUGCAUUUGGUUGUCCUUGGCU |
0.080 |
| 5 |
MIMAT0024577 |
>bta-miR-574 MIMAT0024577
UGAGUGUGUGUGUGUGAGUGUGUG |
0.034 |
| 6 |
MIMAT0017325 |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
0.050 |
| 7 |
MIMAT0004795 |
>hsa-miR-574-5p MIMAT0004795
UGAGUGUGUGUGUGUGAGUGUGU |
0.089 |
| 8 |
MIMAT0005837 |
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA |
0.077 |
| 9 |
MIMAT0005854 |
>mmu-miR-467g MIMAT0005854
UAUACAUACACACACAUAUAU |
0.093 |
| 10 |
MIMAT0022724 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
0.073 |
| 11 |
MIMAT0021390 |
>rmi-miR-5318 MIMAT0021390
UUGUAUAACUGCAGAAGACUUUUCUCC |
0.034 |
| 12 |
MIMAT0017613 |
>aly-miR831-5p MIMAT0017613
AGAAGAGGUACAAGGAGAUGAGA |
0.080 |
| 13 |
MIMAT0032837 |
>cel-miR-8210-5p MIMAT0032837
UGCCUUCUUUCCUUGUGUCGCCGAC |
0.054 |
| 14 |
MIMAT0032835 |
>cel-miR-8209-5p MIMAT0032835
AAAACGAAGAAAGAAGAAGAA |
0.043 |
| 15 |
MIMAT0044920 |
>pab-miR11418 MIMAT0044920
ACCCACCGCCGCUGCCGCCGC |
0.094 |
|
16
|
MIMAT0017325
|
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG
|
0.078
|
|
17
|
MIMAT0022724
|
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU
|
0.055
|
|
18
|
MIMAT0036467
|
>tch-miR-1277-5p MIMAT0036467
UAUAUAUAUAUAUGUACGUAU
|
0.081
|
|
19
|
MIMAT0032837
|
>cel-miR-8210-5p MIMAT0032837
UGCCUUCUUUCCUUGUGUCGCCGAC
|
0.054
|
|
20
|
MIMAT0004885
|
>mmu-miR-467c-5p MIMAT0004885
UAAGUGCGUGCAUGUAUAUGUG
|
0.097
|
|
21
|
MIMAT0009979
|
>hsa-miR-2054 MIMAT0009979
CUGUAAUAUAAAUUUAAUUUAUU
|
0.098
|
|
22
|
MIMAT0005837
|
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA
|
0.097
|
|
23
|
MIMAT0043929
|
>lja-miR11146-3p MIMAT0043929
GCAACGACAUGUAUAGUUGGAGG
|
0.076
|
|
24
|
MIMAT0023986
|
>cgr-miR-598 MIMAT0023986
UACGUCAUCGUCGUCAUCGUUAUC
|
0.057
|
|
25
|
MIMAT0005837
|
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA
|
0.097
|
|
26
|
MIMAT0022724
|
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU
|
0.082
|
|
27
|
MIMAT0036057
|
>chi-miR-211 MIMAT0036057
UUCCCUUUGUCAUCCUUUGCCC
|
0.060
|
|
28
|
MIMAT0000668
|
>mmu-miR-211-5p MIMAT0000668
UUCCCUUUGUCAUCCUUUGCCU
|
0.072
|
|
29
|
MIMAT0012223
|
>dps-miR-980-5p MIMAT0012223
AGUCUCUCACAUGGCUGGUCUAGC
|
0.097
|
|
30
|
MIMAT0022724
|
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU
|
0.070
|
|
31
|
MIMAT0017325
|
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG
|
0.019
|
|
32
|
MIMAT0017325
|
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG
|
0.050
|
|
33
|
MIMAT0024577
|
>bta-miR-574 MIMAT0024577
UGAGUGUGUGUGUGUGAGUGUGUG
|
0.074
|
|
34
|
MIMAT0034619
|
>eca-miR-9074 MIMAT0034619
UGACUAAUAGGAAAUUUUAAGUGAC
|
0.079
|
|
35
|
MIMAT0022724
|
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU
|
0.087
|
|
36
|
MIMAT0001322
|
>ath-miR414 MIMAT0001322
UCAUCUUCAUCAUCAUCGUCA
|
0.029
|
Table 1: Screening the ESTs sequence with lowest E-Value.
To find a novel /unique miRNA 1316 EST sequences were screened for identifying miRNAs of lowest E values. The criterion of lowest E value was between 0.01-0.099. mirbase is an online search engine used to get the micro RNA’s of our specie at the end of screening all the 1316 EST’s only 36 miRNAs were found that meets the criteria of required E value. We blast each EST sequence individually in mirbase and search for values that are of our interest in the transcript Table [9-11]. After getting the required E value we open its accession number and we have its structure and also an option of get sequence we click on it and copy mature RNA sequence and paste that sequence in the word file. The table given below shows the result of screening and also number of A, G, C, U/T, and A-T/U counts are listed in Table 2.
| S. no |
MiRNA |
A Count |
G Count |
C Count |
U/T Count |
Total |
GC Count |
A-T/U Count |
| 1 |
>hsa-miR-6873-5p MIMAT0027646
CAGAGGGAAUACAGAGGGCAAU |
9 |
8 |
3 |
2 |
22 |
11 |
11 |
| 2 |
>bmo-miR-3270 MIMAT0015455
UGUCUUUUUCCUAAAACGAUUCA |
2 |
2 |
5 |
10 |
23 |
7 |
16 |
| 3 |
>hsa-miR-2054 MIMAT0009979
CUGUAAUAUAAAUUUAAUUUAUU |
9 |
1 |
1 |
12 |
23 |
2 |
21 |
| 4 |
>prd-miR-7912-5p MIMAT0030650
CAUUGCAUUUGGUUGUCCUUGGCU |
2 |
6 |
5 |
11 |
24 |
11 |
13 |
| 5 |
>bta-miR-574 MIMAT0024577
UGAGUGUGUGUGUGUGAGUGUGUG |
2 |
12 |
0 |
10 |
24 |
12 |
12 |
| 6 |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
0 |
10 |
0 |
10 |
20 |
10 |
10 |
| 7 |
>hsa-miR-574-5p MIMAT0004795
UGAGUGUGUGUGUGUGAGUGUGU |
2 |
11 |
0 |
10 |
23 |
11 |
12 |
| 8 |
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA |
4 |
8 |
0 |
11 |
23 |
8 |
15 |
| 9 |
>mmu-miR-467g MIMAT0005854
UAUACAUACACACACAUAUAU |
10 |
0 |
5 |
6 |
21 |
5 |
16 |
| 10 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
11 |
2 |
1 |
10 |
24 |
3 |
21 |
| 11 |
>rmi-miR-5318 MIMAT0021390
UUGUAUAACUGCAGAAGACUUUUCUCC |
7 |
4 |
6 |
10 |
27 |
10 |
17 |
| 12 |
>aly-miR831-5p MIMAT0017613
AGAAGAGGUACAAGGAGAUGAGA |
11 |
9 |
1 |
2 |
23 |
10 |
13 |
| 13 |
>cel-miR-8210-5p MIMAT0032837
UGCCUUCUUUCCUUGUGUCGCCGAC |
1 |
5 |
9 |
10 |
25 |
14 |
11 |
| 14 |
>cel-miR-8209-5p MIMAT0032835
AAAACGAAGAAAGAAGAAGAA |
15 |
5 |
1 |
0 |
21 |
6 |
15 |
| 15 |
>pab-miR11418 MIMAT0044920
ACCCACCGCCGCUGCCGCCGC |
2 |
5 |
13 |
1 |
21 |
1 |
3 |
| 16 |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
0 |
10 |
0 |
10 |
20 |
10 |
10 |
| 17 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
11 |
2 |
1 |
10 |
24 |
3 |
21 |
| 18 |
>tch-miR-1277-5p MIMAT0036467
UAUAUAUAUAUAUGUACGUAU |
8 |
2 |
1 |
10 |
21 |
3 |
18 |
| 19 |
>cel-miR-8210-5p MIMAT0032837
UGCCUUCUUUCCUUGUGUCGCCGAC |
1 |
5 |
9 |
10 |
25 |
14 |
11 |
| 20 |
>mmu-miR-467c-5p MIMAT0004885
UAAGUGCGUGCAUGUAUAUGUG |
5 |
7 |
2 |
8 |
22 |
9 |
13 |
| 21 |
>hsa-miR-2054 MIMAT0009979
CUGUAAUAUAAAUUUAAUUUAUU |
9 |
1 |
1 |
12 |
23 |
2 |
21 |
| 22 |
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA |
4 |
8 |
0 |
11 |
23 |
8 |
15 |
| 23 |
>lja-miR11146-3p MIMAT0043929
GCAACGACAUGUAUAGUUGGAGG |
7 |
8 |
3 |
5 |
23 |
11 |
12 |
| 24 |
>cgr-miR-598 MIMAT0023986
UACGUCAUCGUCGUCAUCGUUAUC |
4 |
4 |
7 |
9 |
24 |
11 |
13 |
| 25 |
>mmu-miR-1187 MIMAT0005837
UAUGUGUGUGUGUAUGUGUGUAA |
4 |
8 |
0 |
11 |
23 |
8 |
15 |
| 26 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
11 |
2 |
1 |
10 |
24 |
3 |
21 |
| 27 |
>chi-miR-211 MIMAT0036057
UUCCCUUUGUCAUCCUUUGCCC |
1 |
2 |
9 |
10 |
22 |
11 |
11 |
| 28 |
>mmu-miR-211-5p MIMAT0000668
UUCCCUUUGUCAUCCUUUGCCU |
1 |
2 |
8 |
11 |
22 |
10 |
12 |
| 29 |
>dps-miR-980-5p MIMAT0012223
AGUCUCUCACAUGGCUGGUCUAGC |
4 |
6 |
7 |
7 |
24 |
13 |
11 |
| 30 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
11 |
2 |
1 |
10 |
24 |
3 |
21 |
| 31 |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
0 |
10 |
0 |
10 |
20 |
10 |
10 |
| 32 |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
0 |
10 |
0 |
10 |
20 |
10 |
10 |
| 33 |
>bta-miR-574 MIMAT0024577
UGAGUGUGUGUGUGUGAGUGUGUG |
2 |
12 |
0 |
10 |
24 |
12 |
12 |
| 34 |
>eca-miR-9074 MIMAT0034619
UGACUAAUAGGAAAUUUUAAGUGAC |
10 |
5 |
2 |
8 |
25 |
7 |
18 |
| 35 |
>hsa-miR-1277-5p MIMAT0022724
AAAUAUAUAUAUAUAUGUACGUAU |
11 |
2 |
1 |
10 |
24 |
3 |
21 |
| 36 |
>ath-miR414 MIMAT0001322
UCAUCUUCAUCAUCAUCGUCA |
5 |
1 |
7 |
8 |
21 |
8 |
13 |
Table 2: Screening of different ESTs sequence with count of A, G, C, U/T, and A-T/U.
The structural annotation of these microRNAs to identify their structures through online software called Mfold that uses these sequences to determine the best possible secondary structures and energies of these micro RNA’s. The miRNA structure that is low energy to our required parameters. These 36 miRNA structures are chosen because lowest energy structures are more stable in nature to our required E-value as shown in Supplementary Figure [12].
The functional annotationselect those microRNA whose AU content between 30% and 70%. Only 29% miRNA filled with these criteria in which we found target genes for only 10 sequences. The complete 9 sequences annotated which have basic role in biological process and molecular level is listed in Table 3 [13].
| Peppermint MiRNA |
Function |
GO Term Annotations (biological process) |
GO term annotations (molecular functions) |
>mmu-miR-466i-5p MIMAT0017325
UGUGUGUGUGUGUGUGUGUG |
Protein phosphorylation
electron transport chain
Cell redox homeostasis
Activation of GTPase activity |
Cellular process. GO :0009987 |
Electron transfer activity
GO: 0009055
Phosphotransferase activity
GO: 0016773
GTPase activator activity
GO: 0005096 |
>bmo-miR-3270 MIMAT0015455
UGUCUUUUUCCUAAAACGAUUCA |
Cellular response to nitrogen starvation
Cellular response to fatty acid
Glucosinolate catabolism |
Cellular metabolic process
GO: 0044237 |
ATP binding
GO: 0005524
Ascorbic acid binding
GO: 0031418 |
>hsa-miR-6873-5p MIMAT0027646
CAGAGGGAAUACAGAGGGCAAU |
Somatic cell DNA recombination |
Regulation of biological process
GO: 0065007 |
Sequence specific DNA binding
GO: 0043565 |
>prd-miR-7912-5p MIMAT0030650
CAUUGCAUUUGGUUGUCCUUGGCU |
Defense process
Response to herbivore
Response to fungus |
Response to stimulus
GO: 0050896 |
Not known |
>bta-miR-574 MIMAT0024577
UGAGUGUGUGUGUGUGAGUGUGUG |
Nucleosome assembly |
Cellular component organization
GO: 0071840 |
Histone binding
GO: 0042393 |
>hsa-miR-8485 MIMAT0033692
CACACACACACACACACGUAU |
Intra cellular signal transduction
Transmembrane Protein serine/threonine kinase signaling pathway |
Signaling
GO: 0023052 |
Transmembrane signaling receptor serine threonine kinaseÃÃÃÂ??? activity
GO: 0004674 |
>mmu-miR-467c-5p MIMAT0004885
UAAGUGCGUGCAUGUAUAUGUG |
Regulation of vesicle fusion |
Localization
GO: 0051179 |
Protein dimerization activity
GO: 0046983 |
>rmi-miR-5318 MIMAT0021390
UUGUAUAACUGCAGAAGACUUUUCUCC |
Cell proliferation
Cell differentiation |
Cell proliferation
GO: 0008283 |
2 iron, 2 sulpher cluster binding
GO : 0051537 |
>aly-miR831-5p MIMAT0017613
AGAAGAGGUACAAGGAGAUGAGA |
Double DNA break repair via homologous |
Reproductive process
GO: 0022414 |
Core promoter sequence specific activity
GO : 0001046
mRNA binding
GO: 0003739 |
Table 3: Functional annotation of Mentha piperita with respect 9 different sequence of biological and molecular level.
InterPro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to new protein sequences in order to functionally characterize them. Next step to annotate these sequences which can be done by merging the GOs (obtained from inter pro database) into annotation domain of Blast2Go as shown in Figure 1 [14]. Blast2go helps us to know about our data distribution that how much sequences were annotated with Gene Ontology (GOs). The functional annotation refers to the analysis of the biological processes associated with these components. Those 9 sequences have roles in biological process and at molecular level as well as shown in Figure 2.
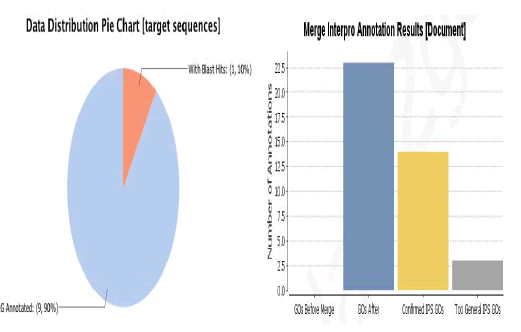
Figure 1: Functional annotation of merge interpro domain reference with gene ontology.
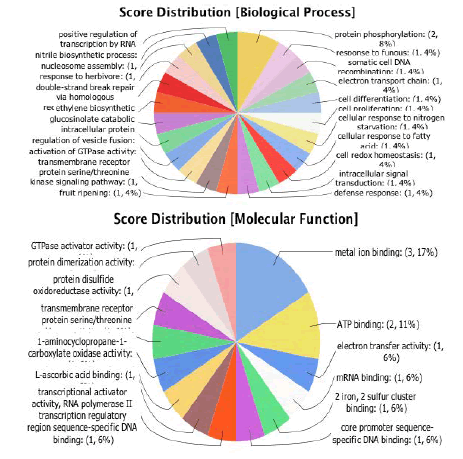
Figure 2: Functional annotation of Mentha piperita with reference to gene ontology biological function, molecular function.
The phylogeny analysis of Mentha has been blasted with Arabidopsis thaliana to confirm the conserved domain by using the online package NCBI database [15]. Based on blast results, the phylogenetic tree was constructed to confirm the conservation of Mentha piperita with Arabidopsis thaliana. The transition and transversion of nucleotide are under observation to have tree and cladogram. The branch length show among the relationship between species as shown in Figure 3.
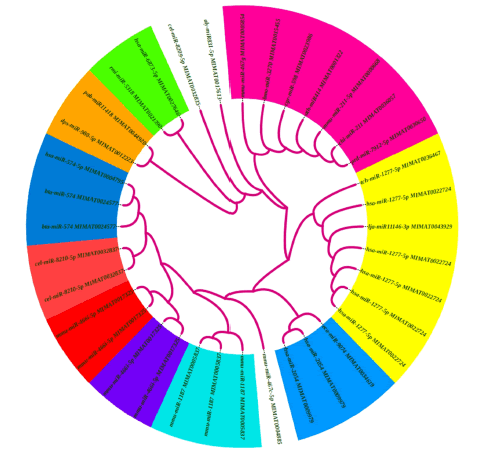
Figure 3: Phylogenetic tree analysis using the neighbor-joining method by MEGA X.
Discussion
Peppermint, commonly known as Mentha piperita, is a perennial herb from the Lamiaceae family [16]. For its medical benefits, including its antispasmodic actions, anti inflammatory and antibacterial peppermint has been used for centuries. Additionally, it is employed in the food and beverage company as a flavoring ingredient. In order to comprehend the molecular processes underlying peppermint's different pharmacological characteristics, recent research has concentrated on locating novel genes and miRNAs in this plant. In this study, we used EST analysis to identify 29 novel genes from peppermint [17]. The functional annotation of these genes revealed their potential roles in various biological processes, including cellular redox homeostasis, electron transport chain, and protein phosphorylation. Furthermore, we identified 36 miRNAs in peppermint, and their functional annotation suggested their involvement in cellular response to nitrogen starvation, glucosinolate catabolism, and somatic cell DNA recombination. The phylogenetic analysis confirmed the conservation of Mentha piperita with Arabidopsis thaliana. In addition, we also calculated the E-values and energies for the identified ESTs and miRNAs, providing further insights into their potential functions. Our findings provide a valuable resource for further studies on the molecular mechanisms underlying the medicinal properties of peppermint. Previous studies have also reported the medicinal properties of peppermint, including its antimicrobial, anti-inflammatory, and antispasmodic effects. Moreover, the identification of novel genes and miRNAs in peppermint has been previously reported, providing insights into the molecular mechanisms underlying its pharmacological properties [18]. Our study builds on these findings, providing novel insights into the potential functions of newly identified genes and miRNAs in peppermint. In this study, we performed transcriptome analysis of Mentha piperita and identified 29 Expressed Sequence Tags (ESTs) using the NCBI database with E-values ranging from 2e-20 to 8e-04. These ESTs were found to be involved in various biological processes including protein phosphorylation, electron transport chain, cell redox homeostasis, activation of GTPase activity, cellular response to nitrogen starvation, cellular response to fatty acid, glucosinolate catabolism, somatic cell DNA recombination, defense process, response to herbivore, response to fungus, nucleosome assembly, cellular component organization, intracellular signal transduction, transmembrane protein serine/threonine kinase signaling pathway, regulation of vesicle fusion, cell proliferation, cell differentiation, double DNA break repair via homologous, and reproductive process. Furthermore, we also identified 36 microRNAs (miRNAs) in Mentha piperita with AU content ranging between 30% and 70%. The energies of these miRNAs were calculated using the online tool RNAfold, with values ranging from -32.1 to -25.6 kcal/mol. The miRNAs were found to have basic roles in various biological processes and molecular functions such as protein phosphorylation, electron transport chain, cellular metabolic process, somatic cell DNA recombination, and regulation of biological process, cellular component organization, signaling, localization, and cell proliferation [19]. To confirm the conservation of Mentha piperita with Arabidopsis thaliana, we performed phylogenetic analysis using the online package NCBI database. The results showed that the transition and transversion of nucleotides were under observation to have tree and cladogram, which confirmed the conservation of Mentha piperita with Arabidopsis thaliana. Overall, our study provides valuable insights into the transcriptome and miRNA profile of Mentha piperita and sheds light on the conservation of this plant with Arabidopsis thaliana. Further investigation into the medicinal properties and prospective uses of plant can be enhanced by the discovery of ESTs and miRNAs involved in various biological processes and molecular functions [20].
Conclusion
In conclusion, this study provides valuable insights into the identification and functional annotation of ESTs and miRNAs in Peppermint (Mentha piperita). The ESTs obtained were identified to be associated with different biological processes, such as stress responses, indicating their potential roles in the growth, metabolism, and development of Peppermint and signal transduction. The miRNAs identified in this study were discovered to have diverse functions regulation of gene expression, modulation of cellular signaling pathways and cellular responses to various stresses. Additionally, the phylogenetic analysis revealed the conservation of Peppermint with Arabidopsis thaliana. The functional annotation of miRNAs revealed that only 29% of the identified miRNAs fulfilled the AU content criteria between 30% and 70%. However, among these miRNAs, 9 sequences were found to have a basic role in molecular functions and biological processes.
Future Recommendations
In future studies, it would be interesting to perform functional validation experiments for the identified ESTs and miRNAs to confirm their roles in peppermint growth and development. Additionally, further investigation of the miRNAs that did not meet the AU content criteria may provide insights into their potential roles in peppermint. The results of this study may provide a foundation for future research aimed at improving the yield and quality of peppermint, which could have potential benefits for the pharmaceutical and food industries.
Data Availability Statement
The data that support the findings were derived from the following resources available in the public domain (GenBank) at https://www.ncbi.nlm.nih.gov/genbank/
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Author Contributions
S.I (Saqib Ishaq), J.K. and R.T designed the study. S.I (Saqib Ishaq), J.K, R.T and O.H performed various in silico analyses. R.T. and S.I wrote the initial MS. A.A, M.I.H, K.G and Y.A, R.N revised the MS. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
The authors acknowledge all those patients who participated in the study.
Funding
This study was financially supported by Department of Computer Sciences and Bioinformatics, Khushal Khan Khattak University, Karak (KKKUK), KPK, Pakistan and Department of Biotechnology and Genetic Engineering, Kohat University of Science and Technology (KUST), Kohat 26000, KPK, Pakistan.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
References
- Bhogireddy S, Mangrauthia SK, Kumar R, Pandey AK, Singh S, et al. (2021) Regulatory non-coding RNAs: A new frontier in regulation of plant biology. Funct Integr Genomics. 21(3):313-330.
[Crossref] [Google Scholar] [PubMed]
- Broughton JP, Lovci MT, Huang JL, Yeo GW, Pasquinelli AE (2016) Pairing beyond the seed supports microRNA targeting specificity. Mol Cell. 64(2):320-333.
[Crossref] [Google Scholar] [PubMed]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 21(18):3674-3676.
[Crossref] [Google Scholar] [PubMed]
- da Silva Ramos R, Rodrigues AB, Farias AL, Simoes RC, Pinheiro MT, et al. (2017) Chemical composition and in vitro antioxidant, cytotoxic, antimicrobial, and larvicidal activities of the essential oil of Mentha piperita L.(Lamiaceae). ScientificWorldJournal. 2017(1):4927214.
[Crossref] [Google Scholar] [PubMed]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. (2009) InterPro: The integrative protein signature database. Nucleic Acids Res. 37(suppl_1):D211-D215.
[Crossref] [Google Scholar] [PubMed]
- Kozomara A, Griffiths-Jones S (2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(D1):D68-D73.
[Crossref] [Google Scholar] [PubMed]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547-1549.
[Crossref] [Google Scholar] [PubMed]
- Li J, Xu C, Wang L, Liang H, Feng W, et al. (2016) PSRna: Prediction of small RNA secondary structures based on reverse complementary folding method. J Bioinform Comput Biol. 14(04):1643001.
[Crossref] [Google Scholar] [PubMed]
- Luo Y, Guo Z, Li L (2013) Evolutionary conservation of microRNA regulatory programs in plant flower development. Dev Biol. 380(2):133-144.
[Crossref] [Google Scholar] [PubMed]
- Mamadalieva NZ, Akramov DK, Ovidi E, Tiezzi A, Nahar L, et al. (2017) Aromatic medicinal plants of the Lamiaceae family from Uzbekistan: Ethnopharmacology, essential oils composition, and biological activities. Medicines. 4(1):8.
[Crossref] [Google Scholar] [PubMed]
- Marzouk MM, Hussein SR, Elkhateeb A, El-Shabrawy M, Abdel-Hameed ES, et al. (2018) Comparative study of Mentha species growing wild in Egypt: LC-ESI-MS analysis and chemosystematic significance. J Appl Pharma Sci. 8(8):116-122.
[Google Scholar]
- McKay DL, Blumberg JB (2006) A review of the bioactivity and potential health benefits of peppermint tea (Mentha piperita L.). Phytother Res. 20(8):619-633.
[Crossref] [Google Scholar] [PubMed]
- Pillai RS, Artus CG, Filipowicz W (2004) Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 10(10):1518-1525.
[Crossref] [Google Scholar] [PubMed]
- Qiu CX, Xie FL, Zhu YY, Guo K, Huang SQ, et al. (2007) Computational identification of microRNAs and their targets in Gossypium hirsutum expressed sequence tags. Gene. 395(1-2):49-61.
[Crossref] [Google Scholar] [PubMed]
- Selvaraj D, Mugunthan SP, Chandra HM, Govindasamy C, Al-Numair KS, et al. (2023) In silico identification of novel and conserved microRNAs and targets in peppermint (Mentha piperita) using expressed sequence tags (ESTs). J King Sau Uni Sci. 35(4):102604.
[Google Scholar]
- Singh P, Pandey AK (2018) Prospective of essential oils of the genus Mentha as biopesticides: A review. Front Plant Sci. 9:1295.
[Crossref] [Google Scholar] [PubMed]
- Synowiec A, Krajewska A (2020) Soil or vermiculite-applied microencapsulated peppermint oil effects on white mustard initial growth and performance. Plants. 9(4):448.
[Crossref] [Google Scholar] [PubMed]
- Tomanic D, Bozin B, Cabarkapa I, Kladar N, Radinovic M, et al. (2022) Chemical composition, antioxidant and antibacterial activity of two different essential oils against mastitis associated pathogens. Acta Veterinaria. 72(1):45-58.
[Google Scholar]
- Vasudevan S (2012) Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 3(3):311-330.
[Crossref] [Google Scholar] [PubMed]
- Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: A small regulatory molecule with big impact. Dev Biol. 289(1):3-16.
[Crossref] [Google Scholar] [PubMed]
Citation: Ishaq S, Khan J, Tahir R, Aziz A, Habib O, et al. (2025) In silico Analysis of MicroRNA of (Mentha X Piperita) Peppermint. Biochem Mol
Biol J. 11:48.
Copyright: �© 2025 Ishaq S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License,
which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.




