Research Article - (2022) Volume 8, Issue 2
Differential Expression of Hirsutella sinensis Genes and Intraspecific Genetic Variation among H. sinensis Strains
Jia-Shi Zhu*
1State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University, China
*Correspondence:
Jia-Shi Zhu, State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University,
China,
Email:
Received: 25-Jan-2022, Manuscript No. IPBMBJ-22-12269;
Editor assigned: 28-Jan-2022, Pre QC No. IPBMBJ-22-12269 (PQ);
Reviewed: 11-Feb-2022, QC No. IPBMBJ-22-12269;
Revised: 18-Feb-2022, Manuscript No. IPBMBJ-22-12269 (R);
Published:
25-Feb-2022, DOI: 10.36648/2471-8084.22.8.58
Abstract
Genetic heterogeneity has been documented among the 17 genotypes of Ophiocordyceps sinensis. However,
intraspecific genetic variations in Hirsutella sinensis (Genotype #1 of O. sinensis) and differential expressions of H.
sinensis genes have rarely been recognized at the genome and transcriptome levels.
Objective: To explore expressions of the H. sinensis genes and intraspecific genetic variations at the genome-
transcriptome levels.
Methods: To cross-analyze GenBank sequences of the assembled genome/mitogenome and transcriptome
assemblies from H. sinensis strains and natural Cordyceps sinensis, and unassembled shotgun genome sequences
and multiple PCR-amplified gene sequences from >300 H. sinensis strains.
Results: Many assembled and unassembled genome sequences were genetically variable and differentially
occurred in the genome assemblies of various H. sinensis strains. Low sequence similarities of some gene transcripts
were also found between the transcriptomic sequences of H. sinensis strain L0106 and natural C. sinensis.
Many genes, including mating-type genes, were differentially transcribed in H. sinensis and natural C. sinensis.
Other genes, including ribosomal 5.8S gene, were transcriptional silencing in H. sinensis and natural C. sinensis.
Multiple genome and transcriptome repeats of numerous genes were identified, some of which contain scattered
nonsense, missense, or frame shift mutant alleles.
Discussion: Differential expressions of H. sinensis genes and apparent intraspecific genetic variations exist among
the H. sinensis strains. Inconsistent occurrence of the mating-type genes in H. sinensis and their paradoxical
transcriptions challenge the hypotheses of homothallism and pseudohomothallism for H. sinensis and suggest
heterothallism. Such genetic and transcriptional variations among H. sinensis strains significantly impact on the
proteomic, chemical, therapeutic and safety profiles of natural C. sinensis and various mycelial fermentation
products that are manufactured using arbitrarily selected strains, warning careful verification of H. sinensis strains prior to academic and industrial/commercial uses.
Introduction
Natural Cordyceps sinensis is a precious therapeutic agent in
traditional Chinese medicine with a rich history of clinical use
for health maintenance, disease amelioration, post-disease recovery,
and antiaging therapy [1-3]. The Chinese Pharmacopoeia
defines natural C. sinensis as an insect-fungi complex, containing
the Ophiocordyceps sinensis fruiting body and a dead
larva from the Hepialidae family [4-6]. Studies over the past
2 decades have demonstrated its heterokaryotic multicellular
structure and dramatic genetic heterogeneity with at least 17
genotypes of O. sinensis and >90 fungal species spanning >37
genera with using various molecular approaches [7-36]. Many
publications on natural C. sinensis and O. sinensis have primarily
focused on Hirsutella sinensis (Genotype #1 and the postulated
anamorph of O. sinensis) through traditional mycology technology
based on fungal morphology and growth characteristics.
However, the observation of Hirsutella-like and H. sinensis-like
morphologies has generated uncertainty among mycologists
in taxonomic determinations of multiple mutant genotypes of O. sinensis and fungi in the families Clavicipitaceae and Ophiocordycipitaceae and in the genera Harposporium and Polycephalomyces [6,12-13,16,30,32,37]. The sequences of AT-biased
genotypes of O. sinensis reside not in the genome of the GC-biased H. sinensis but in the genomes of independent O. sinensis fungi, indicating that the O. sinensis genotypes belong to independent O. sinensis fungi [5-11,14,17-19,26,28,35-36,38].
Molecular marker polymorphism assays and multigene analyses
have demonstrated apparent polymorphic alterations in
multiple H. sinensis strains isolated from C. sinensis specimens
collected from the same or geographically different production
areas [16,39-45]. However, knowledge of intraspecific genetic
variations in Genotype #1 H. sinensis at the genome and transcriptome
levels remains limited.
In ascomycetous fungi, the mating system is usually controlled
by the mating-type (MAT) loci [46]. Bushley et al [24]. and Hu
et al [26]. detected 4 mating-type genes of MAT1-1 and MAT1-2
idiomorphs in the genomes of the H. sinensis strains CS68-2-
1229 and Co18 and hypothesized homothallism/pseudohomothallism
for H. sinensis. Zhang et al [39]. and Zhang and Zhang
[47] found the differential existence of MAT1-1-1 and/or MAT1-
2-1 genes in various H. sinensis strains, hypothesizing facultative
hybridization for H. sinensis for differential occurrence of
(pseudo-) homothallism and heterothallism based on different
genetic materials in various H. sinensis strains. However, these
hypotheses were solely based on genetic evidence without considering
expressions of these H. sinensis mating-type genes.
We explored in this study the differential transcription of H.
sinensis genes and intraspecific genetic variations in H. sinensis through comprehensive cross-analysis of the assembled and
unassembled shotgun genome and mitogenome sequences
from H. sinensis strains, PCR-amplified sequences from multiple H. sinensis strains, transcriptome assemblies from natural C. sinensis and H. sinensis strain L0106. The results of this study
reveal differential transcription of O. sinensis genes (including
the ribosomal 5.8S and mating-type genes) in response to natural
and unnatural conditions and significant intraspecific genetic
variations in the genome and transcriptome sequences of H.
sinensis.
Methods
Gene, Genome, and Mitogenome Sequences of H. Sinensis Strains
Four sets of the assembled shotgun genome sequences,
ANOV00000000, LKHE00000000, LWBQ00000000, and
JAAVMX000000000 of H. sinensis strains Co18, 1229, ZJB12195,
and IOZ07, respectively, and 2 complete mitogenome sequences
(KP835313 of strain 1229 and KY622006 of natural C. sinensis)
are available in GenBank [26,28-29,35-36,48]. (Table S1)
lists 312 H. sinensis strains that were used to obtain assembled
shotgun genome and mitogenome sequences, PCR-amplified
gene sequences, unassembled shotgun genome sequences,
transcriptome shotgun assemblies and a group of mRNA sequences
[4,13,16,24,26,28-29,33,35-36,39,41,49-60].
Sequencing and Assembling Methods for Shotgun
Genome and Mitogenomes Sequences
Genomic DNA from strain Co18 was sequenced with the Roche
454 GS FLX system (Illumina HiSeq: 454), and the shotgun sequences
were assembled using SOAPdenovo v.1.05 and Newbler
v.2.3 under accession #ANOV01000001-ANOV01025873
[26]. Genomic DNA from strain 1229 was sequenced with Illumina
HiSeq sequencing technology, and the shotgun sequences
were assembled using ABySS v.1.2.3 under accession #LKHE01000001-
LKHE01003687 [28]. Genomic DNA from strain
ZJB12195 was sequenced with Illumina sequencing technology
(Hiseq 2000 Sequencing System), and the shotgun sequences
were assembled under accession #LWBQ01000001-LWBQ01000618
using SOAPdenovo v.2.0 [35]. Genomic DNA from
strain IOZ07 was sequenced with PacBio Sequel sequencing
technology, and the shotgun sequences were assembled under
accession #JAAVMX010000001-JAAVMX010000023 using Canu
v.1.7 [36].
The mitochondrial sequences of H. sinensis strain 1229 were
extracted from the filtered reads containing both nuclear and
mitochondrial genomes. The corrected reads were fully assembled
with the Celera Assembler program, refined with Quiver,
and verified by PCR amplification. The mitogenome sequence
KP835313 for strain 1229 is accessible in GenBank [29].
Natural C. sinensis specimens were purchased in Guoluo of Qinghai
Province, China. Total DNA was extracted from the C. sinensis stroma, and randomly sheared to fragments with an average
size of 20 kb for sequencing on a PacBio RS II sequencing platform.
The mitochondrial genome was assembled through Hierarchical
Genome Assembly Process (HGAP) workflow, including
preassembly, error correction, Celera assembly and polishing
with Quiver [61]. The mitogenome sequence KY622006 for natural C. sinensis is accessible in GenBank [57].
The PCR-Amplified Sequences of OSRC14,
OSRC19, OSRC27, And OSRC32 Marker Genes
The OSRC14 marker gene sequences are under the GenBank accession
numbers: KM197544, JQ277381-JQ277382, JQ277386,
JQ277389-JQ277392, JQ325373, JQ325377, JQ325381-
JQ325382, JQ325386, JQ325390, JQ325397-JQ325398,
JQ325402, JQ325408-JQ325409, JQ325422, JQ325429, JQ325431, JQ325438, JQ325442-JQ325443, JQ325451,
JQ325455, JQ325458, JQ325461-JQ325462, JQ325464, and
JQ325472-JQ325487 for 47 H. sinensis strains (Table S1) [39,41-42].
The OSRC19 marker gene sequences of H. sinensis are under
the GenBank accession numbers: JM973741 and JQ277405-
JQ277408 for 5 H. sinensis strains (Table S1) [39,42]).
The OSRC27 marker gene sequences of H. sinensis are under
the GenBank accession numbers: JQ277433-JQ277436,
JQ325605, JQ325609, JQ325613-JQ325614, JQ325618,
JQ325634, JQ325640-JQ325641, JQ325654, JQ325661,
JQ325671, JQ325675, JQ325683, JQ325687, JQ325690,
JQ325693-JQ325694, JQ325696, and JQ325704-JQ325718 for
37 H. sinensis strains (Table S1) [39,41-42].
The OSRC32 marker gene sequences of H. sinensis are under
the GenBank accession numbers: JM973601, JQ277445,
JQ277447-JQ277448, JQ325721, JQ325725-JQ325726,
JQ325729-JQ325730, JQ325734, JQ325750, JQ325756-
JQ325757, JQ325760, JQ325770, JQ325777, JQ325780,
JQ325784, JQ325787, JQ325791-JQ325792, JQ325799,
JQ325803, JQ325806, JQ325809-JQ325810, JQ325812,
JQ325820-JQ325822, JQ325824-JQ325825, JQ325829-
JQ325831, and JQ325833 for 36 H. sinensis strains (Table S1)
[39,41-42].
PCR-amplified Sequences of Other H. sinensis genes
Multiple PCR-amplified H. sinensis sequences in GenBank also
include in this cross-analysis: 116 sequences of MAT1-1-1
gene, 183 sequences of MAT1-2-1 gene, 125 sequences of serine
protease gene (csp1), 45 sequences of beta-tubulin 1 gene
(β-tub1), 51 sequences of translation elongation factor 1-alpha
gene (tef1α), 41 sequences of the largest subunit of RNA polymerase
II (rpb1), 9 sequences of the second largest subunit of
RNA polymerase II (rpb2), as well as partial 18S gene (nrSSU
EF468971) and 28S gene (nrLSU EF468827) sequences of strain
EFCC7287 [4,16,24,26,39,41,47,49-52,54-60].
Shotgun Transcriptome Assemblies of H. sinensis Strain L0106 and Natural C. sinensis and mRNA Sequences of Strain L0106
Two transcriptome assemblies are accessible in GenBank. A
specimen of natural C. sinensis (unknown maturational status)
was collected in Kangding County, Sichuan Province, China. Total
RNA from this specimen was sequenced using 454 technologies.
The sequences longer than 50 bp. from the 454 reads
were assembled into unique sequences (containing contigs
and singletons) using the GS De Novo Assembler software v
2.6 or Newbler 2.6 (454 Life Sciences Corporation, USA). The
shotgun sequences were assembled under GenBank accession
#GAGW01000001-GAGW01016676 using Newbler v.2.3 and
2.6 [53].
The transcriptome assembly GCQL00000000 was derived from
fermented mycelia of strain L0106. The mycelia were collected
for total RNA extraction from cultures grown for 3, 6, and 9
days. Total RNA (20 mg per sample) was subjected to mRNA
purification and total mRNA was used to construct a cDNA
library and sequenced using Illumina HiSeq sequencing technology.
The shotgun nucleotide sequences were assembled
under GenBank accession #GCQL01000001-GCQL01020586
using SOAPdenovo v.2.0 [33]. Additional 41 mRNA sequences
(KP090933-KP090973) from strain L0106 were also sequenced
with Illumina sequencing technology.
Sequence Alignment Analysis
All genome, mitogenome, and transcriptome sequences and
other PCR-amplified DNA sequences were analyzed using the
MegaBlast or discontinuous MegaBlast programs provided by
GenBank (https://blast.ncbi.nlm.nih.gov/).
Results
The ITS1-5.8S-ITS2 Sequences in the Genomes
of H. sinensis Strains
A single copy of ITS1-5.8S-ITS2 sequences was identified in genomes
ANOV00000000, LKHE00000000, and LWBQ00000000
of strains Co18, 1229, and ZJB12195, respectively, using the
2nd generation of sequencing technology but multiple copies
of ITS1-5.8S-ITS2 sequences in genome JAAVMX000000000 of
strain IOZ07 with using the 3rd generation of sequencing technology
(Table 1) [26,28,35-36]. These H. sinensis ITS sequences are GC-biased and 80.1%-89.9% similar to the sequences of
AT-biased Genotypes #4-6, #15-17 of O. sinensis.
Table 1: Comparisons of the ribosomal ITS1-5.8S-ITS2 sequences of H. sinensis strains.
| H. sinensis strain |
ITS1-5.8S-ITS2 sequence |
% Similarity to GC-biased AB067721 (59→549) of strain GYOKUJU |
| Accession # |
Range & direction |
| 1229 |
LKHE01000582 |
2,132→2,622 |
100% (491/491) |
| Co18 |
ANOV01021709 |
896→1,386 |
99.8% (490/491) |
| ZJB12195 |
LWBQ01000008 |
991,797→992,287 |
99.4% (488/491) |
| IOZ07 |
JAAVMX010000002 |
18,688,917→18,689,407 |
100% (491/491) |
| 18,702,095→18,702,586 |
97.4% (485/498) |
| JAAVMX010000008 |
13,823→14,313 |
100% (491/491) |
| 1,199→1,687 |
99.2% (488/492) |
| JAAVMX010000017 |
9,147←9,637 |
100% (491/491) |
| 21,791←22,281 |
100% (491/491) |
| 34,435←34,925 |
100% (491/491) |
| 47,079←47,569 |
100% (491/491) |
| JAAVMX010000018 |
13,381→13,871 |
100% (491/491) |
| 26,076→26,566 |
100% (491/491) |
| 38,771→39,261 |
100% (491/491) |
| 51,467→51,958 |
99.0% (488/493) |
| 700→1,186 |
97.0% (479/494) |
| JAAVMX010000019 |
19,404→19,894 |
100% (491/491) |
| 32,048→32,538 |
100% (491/491) |
| 6,233→6,733 |
95.5% (476/498) |
| 44,729→45,251 |
91.8% (480/523) |
(Figure 1) shows illustratively the locations of the H. sinensis ITS1-5.8S-ITS2 (59→549 of AB067721; 896→1,386
of ANOV01021709; 2,132→2,622 of LKHE01000582;
991,797→992,287 of LWBQ01000008; multiple segments
of JAAVMX000000000) and several partial 18S and 28S
gene sequences of strains GYOKUJU, Co18, 1229, ZJB12195,
IOZ07, EFCC7287, and YN07-8. LKHE01000582 (dark
blue), LWBQ01000008, ANOV01021709/ANOV01022831/
ANOV01024581, and JAAVMX010000002/JAAVMX010000008/
JAAVMX010000017-JAAVMX010000019 (light blue) are the
assembled genome sequences from H. sinensis strains 1229,
ZJB12195, Co18, and IOZ07, respectively [26,28,35-36].
AB067721 (red) is the PCR-amplified ITS1-5.8S-ITS2 sequence
of strain GYOKUJU [13]. EF468971 and EF468827 (green) are
the PCR-amplified partial 18S and 28S genes sequences, respectively,
from strain EFCC7287 [4]. JM973574-JM973579
(brown) are unassembled shotgun partial 28S gene sequences
of strain YN07-8 [39].
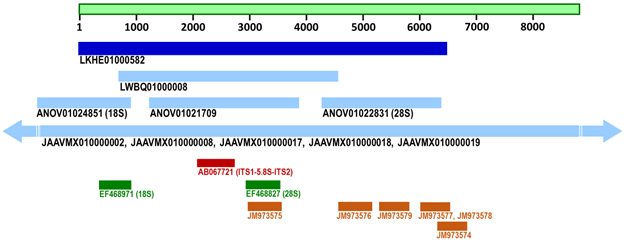
Figure 1: Illustration of the locations of the genome segments of nuclear ribosomal DNA relative to the genome assembly sequence LKHE01000582.
The 5.8S Gene Sequence of H. sinensis Strains
The ribosomal 5.8S gene sequence (218→373 of AB067721
of strain GYOKUJU) is 100% homologous to most of the 5.8S
genome sequences of strains Co18, 1229, ZJB12195 and
IOZ07 [13,26,28,35-36]. However, several copies of the 5.8S
gene of strain IOZ07 contain some mutant alleles (Figure
S1): 18,702,254→18,702,409 (97.5%) of JAAVMX010000002,
858→1,011 (95.6%) of JAAVMX010000018, and 6,396→6,550
and 44,889→45,046 (95.6% and 98.7%) of JAAVMX010000019.
The mismatched alleles may cause translational interruptions
of the once-functional 5.8S gene due to nonsense, frame shift,
or missense allelic mismatches. All the 5.8S gene sequences of
GC-biased H. sinensis (Genotype #1 of O. sinensis) are 79.8%-
89.6% similar to the 5.8S gene sequences of the AT-biased
Genotypes #4-6, #15-17 of O. sinensis [6,9,26,28,35-36]. No
additional genome sequences of strains Co18, 1229, ZJB12195,
and IOZ07 show >89.9% similarities with any of the AT-biased
5.8S gene sequences, comfirming that the sequences of the
AT-biased genotypes of O. sinensis belong to the genomes
of independent fungi [6,8-11]. The transcriptome assembly GAGW00000000 of natural C. sinensis contains no 5.8S gene
sequence [53].
The 18S Gene Sequence of LKHE01000582 of H.
sinensis Strain 1229
The ribosomal 18S gene segment 1→2,132 of LKHE01000582
of strain 1229 is 99.9%-100% homologous to the non-overlapped
sequences ANOV01024851 (427→1,198) and
ANOV01021709 (1→927) of strain Co18 and LWBQ01000008
(990,360→991,828) of strain ZJB12195 and 100% homologous
to multiple segments of the genome assembly
JAAVMX000000000 of strain IOZ07: 18,686,785→18,688,948 of
JAAVMX010000002; 990,360→991,828 of JAAVMX010000008;
9,606→11,768, 22,250→24,412, 34,894→37,056,
47,538→49,700 of JAAVMX010000017; 11,250→13,412,
23,945→26,107, 36,640→38,802 of JAAVMX010000018;
and 4,095→6,233, 17,273→19,435, 29,917→32,079 of
JAAVMX010000019 [26,28,35-36]. However, the 18S gene
sequence of LKHE01000582 is 97.1%-98.9% similar to 5
other 18S gene segments: 18,699,944→18,702,095 of
JAAVMX010000002, 1→1,199 of JAAVMX010000008, 1→730
of JAAVMX010000018, and 4,095→6,265 and 42,577→44,760
of JAAVMX010000019, with scattered transition, transversion
and insertion/deletion mutant alleles, which may lead to nonsense,
frame shift, or missense mutations of the 18S gene.
The PCR amplified partial 18S gene sequence (EF468971)
of H. sinensis strain EFCC7287 is 99.4%-99.8% homologous
to 18S sequences JX968024-JX968028 obtained
from various H. sinensis strains using the same pair of
primers. EF468971 is >98.6% homologous to the assembled genome sequences ANOV01024581, LKHE01000582,
LWBQ01000008, JAAVMX010000002, JAAVMX010000008, and
JAAVMX010000017-JAAVMX010000019 of strains Co18, 1229,
ZJB12195, and IOZ07, respectively [4,26,28,35-36].
The 18S gene segment (1→2,132) of LKHE01000582 of H. sinensis strain 1229 is 99.0%-100% homologous to multiple transcriptome
sequences of natural C. sinensis: GAGW01005077/
GAGW01005078/GAGW01005953/GAGW01012978/
GAGW01013875/GAGW01016121/GAGW01013937 [28,53].
The longest transcript GAGW01005077 overlaps with other 6
transcripts.
The 18S gene sequence EF468971 of strain EFCC7287 is 98.6%-
99.8% homologous to the overlapped or partially overlapped
transcriptome sequences: GAGW01005077/GAGW01005078/
GAGW01013937 of natural C. sinensis, indicating that the transcripts
might be derived from independent fungi that co-colonized
in natural C. sinensis [4,53].
The 28S Gene Sequence of LKHE01000582 of H.
sinensis Strain 1229
The ribosomal 28S gene segment 2,623→6,454 of
LKHE01000582 of strain 1229 is 82.2%-100% similar to multiple
28S genome segments: ANOV01021709 and ANOV01022831;
LWBQ01000008; and JAAVMX010000002/JAAVMX010000008/
JAAVMX010000017-JAAVMX010000019 of strains Co18, 1229,
and IOZ07, respectively [26,28,35-36], containing numerous,
scattered insertion/deletion mutations and some transition
and transversion mutant alleles, some of which may cause
nonsense, frame shift, or missense mutations of the 28S genes
(Table 2).
Table 2: Comparisons of the ribosomal 28S sequences of H. sinensis strains
| Strain |
28S gene sequence |
% Similarity vs. LKHE01000582 (2,623→6,454) of strain 1229 |
| Accession # |
Range & direction |
| Co18 |
ANOV01021709 |
1,387→2,626 |
100% (1,240/1,240) |
Not overlapped |
| ANOV01022831 |
1→2,057 |
95.6% (2,018/2,110) |
| ZJB12195 |
LWBQ01000008 |
995,230→996,749 |
98.7% (1,507/1,527) |
|
| 992,288→994,347 |
96.9% (1,996/2,060) |
|
| 93,757→94,224 |
82.2% (447/544) |
|
| IOZ07 |
JAAVMX010000002 |
18,689,408→18,693,238 |
99.8% (3,828/3,832) |
|
| 18,702,587→18,706,423 |
98.9% (3,807/3,851) |
|
| JAAVMX010000008 |
14,314→18,144 |
99.8% (3,828/3,832) |
|
| 1,688→5,516 |
99.5% (3,817/3,838) |
|
| JAAVMX010000017 |
5,316←9,146 |
99.8% (3,828/3,832) |
|
| 17,960←21,790 |
99.8% (3,828/3,832) |
|
| 30,604←34,434 |
99.8% (3,828/3,832) |
|
| 43,248←47,078 |
99.8% (3,828/3,832) |
|
| 55,943←58,385 |
99.8% (2,440/2,445) |
|
| JAAVMX010000018 |
13,872→17,702 |
99.8% (3,828/3,832) |
|
| 26,567→30,397 |
99.8% (3,828/3,832) |
|
| 39,262→43,092 |
99.8% (3,828/3,832) |
|
| 1,188→5,007 |
99.3% (3,808/3,836) |
|
| 51,959→55,162 |
98.4% (3,168/3,218) |
|
| JAAVMX010000019 |
19,895→23,725 |
99.8% (3,828/3,832) |
|
| 32,539→36,371 |
99.8% (3,827/3,835) |
|
| 6,734→10,576 |
98.8% (3,810/3,858) |
|
| 45,253→49,110 |
98.1% (3,804/3,877) |
|
Cross-analysis revealed that the 28S genome sequence
ANOV01022831 of H. sinensis strain Co18 is 92.9%-95.7% similar
to the genome segments of LKHE01000582/LKHE01002349,
JAAVMX010000002/JAAVMX010000008/JAAVMX010000017-
JAAVMX010000019, and LWBQ01000008/LWBQ01000038 of
strains 1229, IOZ07, and ZJB12195 with multiple insertions/
deletions and transition and transversion point mutations
[26,28,35-36].
The PCR-amplified partial 28S RNA gene sequence EF468827
of H. sinensis strain EFCC7287 is 99.5%-99.8% homologous to
other 28S gene sequences JX968029-JX968033 of various H.
sinensis strains and the genome sequences ANOV01021709,
LKHE01000582, LWBQ01000008, and JAAVMX010000002/
JAAVMX010000008/JAAVMX010000017-JAAVMX010000019
of strains Co18, 1229, ZJB12195, and IOZ07, respectively
[4,26,28,35-36].
Of the 254 unassembled shotgun H. sinensis sequences
(JM973567-JM973820) of H. sinensis strain YN07-8, JM973574-
JM973579 are partial 28S gene sequences at various locations
(Figure 1) [39]. JM973577 and JM973578 are overlapped partially
overlapped with JM973574.
JM973575 of strain YN07-8 is 99.5%-99.8% homologous to the
overlapped sequence EF468827 of H. sinensis strain EFCC7287
and genome sequence ANOV01021709 of train Co18 [4,26,39].
JM973574 and JM973576-JM973579 locate downstream
of LWBQ01000008 and do not align with any part of the genome
sequence LWBQ00000000 (Figure 1) [35,39]. JM973574
and JM973577-JM973578 of strain YN07-8 are 98.7%-100%
homologous to the genome segments of ANOV01022831,
LKHE01000582, and JAAVMX010000002/JAAVMX010000008/
JAAVMX010000017-JAAVMX010000019 of strain Co18, 1229,
and IOZ07, respectively [26,28,36,39]. JM973576 and JM973579
of strain YN07-8 are 99.3%-99.7% homologous to segments of
JAAVMX010000002/JAAVMX010000008/JAAVMX010000017-
JAAVMX010000019 and LKHE01000582 of strains IOZ07 and
1229 but only 95.3% and 86.2% similar to ANOV01022831 of
strain Co18 with scattered insertion/deletion, transition, and
transversion mutant alleles (Figure S2).
The 28S gene segment (2,623→6,454) of LKHE01000582 of H. sinensis strain 1229 is 97.6%-100% homologous to at least
23 segments of the transcriptome assembly GAGW00000000
of natural C. sinensis, but <97% similar to at least 11 other
GAGW00000000 segments with scattered transition, transversion,
and insertion/deletion, mutant alleles (Table S2) [28,53].
Some of these transcriptome sequences are overlapped or partially
overlapped, likely indicating divergent genome sources of
multiple fungi co-colonized in natural C. sinensis.
The overlapped 28S gene sequences EF468827 and JM973575
of strains EFCC7287 and YN07-8 are 99.4%-100% homologous
to the transcriptome sequences GAGW01000468/
GAGW01000467/GAGW01005959/GAGW01013228/
GAGW01014446 of natural C. sinensis, some of which are overlapped
[4,39,53].
The partial 28S gene sequences JM973574, JM973576-
JM973579 of strain YN07-8 are 99.4%-100% homologous to
transcriptome sequence GAGW01000465 but 78.5%-100%
similar to other 6-10 overlapped transcriptome sequences of natural C. sinensis [39,53].
Cross-analysis revealed that the transcriptome sequence
1→720 of GAGW01000465 of natural C. sinensis is 99.4%-
100% homologous to the genome sequences of LKHE01000582
(4,282→5,001), ANOV01022831 (5→724); LWBQ01000008
(overlapped 994,000→994,347 and 996,402→996,749) of
strains 1229, Co18, and ZJB12195, and numerous segment
sequences of strain IOZ07: 18,691,067→18,691,786 and
18,704,249→18,704,967 of JAAVMX010000002; 3,349→4,068;
and 15,973→16,692 of JAAVMX010000008; 6,768←7,487;
19,412←20,131; 32,056←32,775; 44,700←45,419; and
57,395←58,113 of JAAVMX010000017; 2,836→3,555;
15,531→16,250; 28,226→28,945; 40,921→41,640; and
53,621→54,339 of JAAVMX010000018; and 8,410→9,128;
21,554→22,273; 34,198→34,917; and 46,925→47,644 of
JAAVMX010000019 [26,28,36,53].
Second part (720→1,682) of the transcriptome sequence
GAGW01000465 is 99.3%-99.5% homologous to the genome
sequences of LKHE01000582 (5,341→6,306) of strain
1229 and numerous segment sequences of strain IOZ07:
18,692,126→18,693,091 of JAAVMX010000002; 4,406→5,369
and 17,032→17,997 of JAAVMX010000008; 5,463←6,428;
18,107←19,072; 30,751←31,716; 43,395←44,360; and
56,090←57,055 of JAAVMX010000017; 3,895→4,860;
16,590→17,555; 29,285→30,250; 41,980→42,945; and
54,678→55,162 of JAAVMX010000018; and 9,465→10,430;
22,613→23,578; 35,259→36,223; and 47,988→48,952 of
JAAVMX010000019 [26,28,36,53]. Slightly lower similarities
(97.8%-98.1%) were also found between the transcriptome
segment 720→1,682 of GAGW01000465 and genome sequences:
18,705,306→18,706,280 of JAAVMX010000002,
54,678→55,162 of JAAVMX010000018, and 47,988→48,952
of JAAVMX010000019 with scattered insertion/deletion point
mutant alleles and a few transition and transversion point mutations
[36,53]. This segment of GAGW01000465 is only 93.5%
similar to the genome sequence ANOV01022831 (5→724) of
strain Co18 with multiple mismatched alleles [26,53].
Mating-Type Genes of H. sinensis
Table 3 lists 235 H. sinensis strains that contain either or both
MAT1-1-1 and MAT1-2-1 genes listed in GenBank [24,26,28,35-36,40,62]. Twenty two of the strains contain only MAT1-1-1
gene but no MAT1-2-1 gene; 63 contain only MAT1-2-1 gene
but no MAT1-1-1 gene; and 150 contain both MAT1-1-1 and
MAT1-2-1 genes.
Table 3: H. sinensis strains contain either or both MAT1-1-1 and MAT1-2-1 genes listed in GenBank.
| Containing only MAT1-1-1 gene (N=12) |
Containing only MAT1-2-1 gene (N=30 |
Containing both MAT1-1-1 & MAT1-2-1 genes (N=55) |
| CS09-143 |
CS09-225 |
1229 |
SC04 |
| CS09-229 |
CS26-277 |
Co18 |
SC05 |
| CS68-2-1228 |
CS34-291 |
CS09-111 |
SC06 |
| GS03 |
CS36-1294 |
CS09-121 |
SC07 |
| IOZ07 |
CS37-295 |
CS18-266 |
SC08 |
| SC08 |
CS70-1211 |
CS2 |
SC09_65 |
| SC09_97 |
CS71-1220 |
CS25-273 |
SC09_200 |
| XZ05_6 |
ID10_1 |
CS560-961 |
TB01 |
| XZ06_260 |
NP10_1 |
CS561-964 |
TB02 |
| XZ07_H2 |
QH02 |
CS6-251 |
TB03 |
| XZ09_95 |
QH05 |
CS68-2-1229 |
TB04 |
| YN09_61 |
QH09_111 |
CS68-5-1216 |
TB05 |
|
QH09_157 |
CS70-1208 |
TB06 |
|
QH-YS-199 |
CS71-1218 |
TB07 |
|
SC09_47 |
CS71-1219 |
TB08 |
|
SC09_57 |
CS76-1284 |
XZ05_2 |
|
SC09_77 |
CS91-1291 |
XZ05_8 |
|
SC-3 |
GS01 |
XZ12_16 |
|
SC-5 |
GS02 |
YN01 |
|
XZ06_124 |
GS04 |
YN02 |
|
XZ-LZ06_1 |
GS05 |
YN03 |
|
XZ-LZ06_108 |
QH01 |
YN09_3 |
|
XZ-LZ07_H1 |
QH03 |
YN09_22 |
|
XZ-NQ_154 |
QH04 |
YN09_51 |
|
XZ-NQ_180 |
QH06 |
YN09_64 |
|
XZ-SN_44 |
QH07 |
|
|
YN09_6 |
QH08 |
|
|
YN-1 |
SC01 |
|
|
YN-4 |
SC02 |
|
|
ZJB12195 |
SC03 |
|
Bushley et al [24]. detected 3 mating-type genes of MAT1-1 idiomorph
in the genome sequence KC437356 of strain CS68-2-
1229: MAT1-1-1 (6,530→7,748), MAT1-1-2 (4,683→6,183), and
MAT1-1-3 (3,730→4,432). These genes are 99.9%-100% homologous
to the genome assemblies: LKHE01001116 (3,691←4,909;
5,374←6,874; 7,125←7,827), JAAVMX010000001
(6,698,911→6,700,129; 6,696,939→6,698,439;
6,695,986→6,696,688), and ANOV01017390 (302←1,519) and
ANOV01017391 (276←1,776; 2,027←2,729) of strains 1229,
IOZ07, and Co18, respectively, but absent from the genome
assembly LWBQ0000000 of strain ZJB12195 [24,26,28,35-36].
The MAT1-1-1 sequence of KC437356 is 98.4%-100% homologous
to 34 MAT1-1-1 gene sequences of H. sinensis in Gen-
Bank, but no other MAT1-1-2 or MAT1-1-3 gene sequence of H.
sinensis listed in GenBank.
The MAT1-1-1, MAT1-1-2, and MAT1-1-3 sequences of
KC437356 of strain CS68-2-1229 are absent from the transcriptome
assembly GCQL00000000 of H. sinensis strain L0106
[24,33]. The MAT1-1-1 sequence is 94.2%-94.6% similar to the
transcriptome segment (297←1,129) GAGW01008880 of natural C. sinensis with a 48-nt. deletion between nucleotides 358
and 359 of GAGW01008880 [53].
The MAT1-2-1 sequence JQ325153 of H. sinensis strain
GS09_121 is 99.7%-99.9% homologous to the sequences of
the genome assemblies LWBQ01000021 (238,864←239,736),
LKHE01001605 (13,851←14,723), and ANOV01000063
(9,319→10,191) of strains Co18, 1229 and ZJB12195, respectively,
but absent from the genome assembly JAAVMX000000000
of strain IOZ07 [24,26,28,35-36,42]. JQ325153 is 97.3%-100%
homologous to 85 other MAT1-2-1 sequences of H. sinensis strains (Table 3).
JQ325153 is 99.6% homologous to segment 388←671 of
the transcriptome assembly GCQL01020543 of strain L0106
but 90.3% similar to segment 672←1,153 of GCQL01020543
with a 52-nt. deletion [24,33]. However, the sequence of the
MAT1-2-1 gene was absent from the transcriptome assembly
GAGW00000000 of natural C. sinensis [53].
Translation Elongation Factor 1α (tef1α) Gene
Sequence hologogies of 98.8%-100% were found among the
translation elongation factor 1α (tef1α) genes of 52 H. sinensis strains [4,42,50,52,54-58]. The tef1α gene was found less sensitive
in fungal species identification, because the sequence of H. sinensis tef1α gene is 96.2%-96.9% similar to the sequences
of O. robertsii (EF468766; KC561979), O. karstii (KU854945;
KU854946), O. lanpingensis (KC417462; KC417463), and other
Hirsutella sp. (KY415601).
The tef1α sequence EF468767 of strain EFCC7287 is 99.5%-
99.7% homologous to 4 overlapped or partially overlapped transcriptome
sequences of natural C. sinensis: GAGW01000517
(332←1,005), GAGW01003987 (1←263), GAGW01014172
(250←479), and GAGW01013074 (1→212) [4,53]. It is absent
from the transcriptome assembly GCQL00000000 of the H. sinensis strain L0106 [4,33], indicating that tef1α gene was in a
silent status in H. sinensis. EF468767 and its longest transcriptome
sequence GAGW01000517 are 99.4%-99.8% homologous
to a single gene copy (JAAVMX010000011, ANOV01000106,
LKHE01001641, and LWBQ01000064) of each of the genome
assemblies of strains IOZ07, Co18, 1229, and ZJB12195, respectively
[4,26,28,35-36,53]. Some of the overlapped tef1α transcripts
may likely be derived from various fungi co-colonized in
natural C. sinensis.
The Largest and Second Largest Subunits of
RNA Polymerase ІІ (rpb1 and rpb2) Genes
Sequence hologogies of 98.8%-100% were found among the
largest subunit of RNA polymerase ІІ (rpb1) genes of 41 H.
sinensis strains [4,42,52,54-56]. EF468874 (rpb1) of H. sinensis strain EFCC7287 is 100% homologous to a single gene
copy (ANOV010001113, LKHE01001285, LWBQ01000001,
and JAAVMX010000003) of each of the genome sequences
of strains Co18, 1229, ZJB12195, and IOZ07, respectively
[4,26,28,35-36]. EF468874 is 99.0%-100% to the transcriptome
sequences GCQL01000113 of H. sinensis strain L0106 and
GAGW01009638 of natural C. sinensis [4,33,53].
Sequence hologogies of 99.4%-100% were found among the
second largest subunit of RNA polymerase ІІ (rpb2) genes of
9 H. sinensis strains [4,42,52,54-56]. EF468924 (rpb2) of H.
sinensis strain EFCC7287 is 99.4% homologous to the genome
sequences ANOV010007657, LKHE01001069, LWBQ01000010,
and JAAVMX010000012 of strains Co18, 1229, ZJB12195, and
IOZ07, respectively, but 94.3% similar to another ZJB12195
sequence LWBQ01000044 with a 39-nt. insertion [4,26,28,35-36]. EF468924 is 97.6%-100% homologous to the transcriptome
sequences GCQL01011291 of H. sinensis strain
L0106 and 3 non-overlapped transcripts (GAGW01012703/
GAGW01015334/GAGW01001851) of natural C. sinensis [4,33,53].
Serine Protease (csp1) Gene
Sequence hologogies of 96.5%-100% were found among the serine
protease (csp1) genes of 125 H. sinensis strains [15,39,42,54].
JQ325256 (csp1) of strain GS09-225 is 99.0%-100% homologous
to the genome sequences ANOV0100009487, LKHE01000343,
LWBQ01000085, JAAVMX010000012 and JAAVMX010000021
of strains Co18, 1229, ZJB12195, and IOZ07, respectively
[4,26,28,35-36]. JQ325256 is 65.9%-70.0% similar to the
overlapped transcriptome sequences GCQL01005668 and
GCQL01005996 of H. sinensis strain L0106 but absent from the
transcriptome assembly GAGW00000000 of natural C. sinensis [4,33,53], indicating transcriptional silencing of csp1 gene in
strain L0106 and natural C. sinensis.
Beta-Tubulin 1 (β-tub1) Gene
Sequence hologogies of 96.9%-100% were found among
the beta-tubulin 1 (β-tub1) genes of 45 H. sinensis strains
[15,39,42,54]. JX968019 (β-tub1) of strain QH09-201 is 99.4%-
99.8% homologous to the genome assemblies ANOV01001731,
LKHE01000036, LWBQ01000017/LWBQ01000184, and
JAAVMX010000003 of strains Co18, 1229, ZJB12195, and
IOZ07, respectively [26,28,35-36]. JX968019 is 100% homologous
to 3 segments (1→312; 311→613; 611→838) of the transcriptome
assembly GAGW01002303 of natural C. sinensis and
99.0% to a short transcript (1←197) of GCQL01016424 (241
bp) of strain L0106 [33,42,53].
Mitogenome Sequences of H. sinensis and Natural C. sinensis
Complete mitogenome sequence KP835313 of H. sinensis strain
1229 is 99.9% (157,454/157,584) homologous to KY622006 of
natural C. sinensis with 119 gaps (up to 8-base) of insertions/
deletions, but 11 pairs of the short mitogenome segments of
KP835313 and KY622006 share 84.9%-98.3% similarities with
multiple transition, transversion, and insertion/deletion point
mutations [33,48].
Blasting KP835313 of H. sinensis strain 1229 against the assembled
genome sequences in GenBank database revealed best
hits on 217 subject sequences of strain Co18, 1229, ZJB12195
and IOZ07, 138 of which are highly homologous (97.1%-100%)
to KP835313, but 79 are <97% similar to KP835313 with scattered
transition, transversion and insertion/deletion mutant
alleles [26,28-29,35-36].
Of the 254 unassembled shotgun genome sequences of strain
YN07-8, sequences JM973570-JM973573, and JM973767 share
99.4%-99.9% homologies with the mitogenomic sequence
KP835313 [29,39].
Blasting the mitogenome sequence KP835313 against the
assembled transcriptome sequences hits on 1,455 O. sinensis transcriptome sequences, many of which are overlapped
[29,33,53]. KP835313 is 96.8%-100% homologous to 250
best-hit transcriptome sequences, 198 (ranging 496-4820
nt. in length) of which are segments of the transcriptome
GAGW00000000 of natural C. sinensis and the rest 52 (ranging
533-3227 nt. in length) are segments of the transcriptome
GCQL00000000 of strain L0106.
Blasting the segment 5,261→5,451 of KP835313 hits on 85
overlapped GAGW00000000 transcripts of natural C. sinensis that cover >90% of the segment length of KP835313, or 101
transcript repeats that cover >80% of the segment length of
KP835313 [33,53]. These GAGW00000000 transcripts share
94.4%-100% similarities with KP835313 sequences with scattered
insertion/deletion, transition and transversion mutant
alleles in some of the repeats, indicating the GAGW00000000
transcript repeats might likely be derived from multiple fungi
co-colonized in natural C. sinensis.
Another mitogenome segment 1→506 of KP835313 shares
99.4% homology with mitochondrial RNA ligase gene transcript
GAGW01012749 of natural C. sinensis [33,53]. GAGW01012749
is 97%-100% homologous to 64 overlapped GAGW00000000
transcripts that cover ≥ 90% of the length of GAGW01012749,
or to 122 transcript repeats that cover ≥ 80% of the length of
GAGW01012749 with scattered insertion/deletion and transition
and transversion mutant alleles.
Both segments (1→506 and 5,261→5,451) of KP835313 did not
align to any part of the transcriptome assembly GCQL00000000
of strain L0106 [29,33].
Genome Sequence ANOV01021101 of H. sinensis Strain Co18
Segment 1→2,009 of ANOV01021101 of strain Co18 is 97.1%
similar to segments 22,379→24,387 of LKHE01000642 and
435,899→437,907 of JAAVMX010000012 of strains 1229 and
IOZ07, respectively (Figure S3), but absent from the genome
assembly LWBQ00000000 of strain ZJB12195 [26,28,35-36].
Cross-analysis showed that LKHE01000642 is 97.1%-100% homologous
to segments of JAAVMX010000012, LWBQ01000085,
ANOV01002198/ANOV01021101/ANOV01021102 of strains
IOZ07, ZJB12195, and Co18, respectively.
Segment 368→2,009 of ANOV01021101 is 99.9% homologous
to segment 80→1,721 of the transcriptome sequence
GCQL01010475 of strain L0106 [26,33]. Segment 24→638 of
ANOV01021102 of strain Co18 is 100% homologous to overlapped
transcripts GCQL01014530 (1←649) and GCQL01010475
(1,873→2,487) of strain L0106 [26,33]. ANOV01021101 and
ANOV01021102 did not align to any part of the transcriptome
assembly GAGW00000000 of natural C. sinensis [26,53]. These
results indicate transcriptional silencing of the genes of all fungi
co-colonized in natural C. sinensis and transcriptional activation
of the genes in strain L0106.
Genome Sequence LKHE01000676 of H. sinensis Strain 1229
The sequence LKHE01000676 of strain 1229 is 97.0%-100% homologous
to the multiple genome sequences ANOV01000288/
A N O V 0 1 0 0 0 2 8 9 /A N O V 0 1 0 0 0 8 1 1 /A N O V 0 1 0 0 0 8 1 2 /
A N O V 0 1 0 0 1 6 7 6 /A N O V 0 1 0 0 7 1 5 9 /A N O V 0 1 0 0 9 8 7 6 /
ANOV01009877/ANOV01022491, JAAVMX010000004, and
LWBQ01000084, but shares low similarities with many other
segment sequences of the genome assemblies ANOV00000000,
JAAVMX000000000, and LWBQ00000000 of strains Co18,
IOZ07, and ZJB12195, respectively (Table S3). For instance, segment 827→1,351 of LKHE01000676 is 82.0%-91.9% similar
to segments of ANOV01006005, JAAVMX010000001/
JAAVMX010000004, LWBQ01000135, and many other segments
of strains Co18, IOZ07, and ZJB12195, respectively
[26,28,35-36]. Figure 2 show sequence comparisons with scattered
transition and transversion mutations and multiple insertion/ deletion mutant alelles.

Figure 2: Alignment of the genome assembly segments LKHE01000676 with other genomic sequences.
Cross-analysis revealed that the lengthy segments 4,337→9,693
and 11,500→18,158 of ANOV01000289 of strain Co18 are 91.0%-
96.5% similar to segments 6,005←12,651 and 15,974←21,281
of LKHE01000676; 5,342←11,988 and 15,311←20,620 of
JAAVMX010000004 of strains 1229 and IOZ07, respectively, with scattered transition, transversion, insertion/deletion
mutant alleles [26,28,36]. Within these lengthy segments
of ANOV01000289, 2 shorter segments 5,485→6,702 and
16,257→17,663 are 99.2%-99.9% homologous to the segments
of LWBQ01000084 of strain ZJB12195 [26,35].
Segment 4,490→4,881 of ANOV01006005 of strain Co18
is 97.7% homologous to segment 4,011,040→4,011,426
of JAAVMX010000001 and segment 52,870→53,256 of
LKHE01002757 of strains IOZ07 and 1229, respectively
[26,28,35-36]. This segment of ANOV01006005 is only 91.8%
similar to segment 350,593→351,007 of LWBQ01000080 of
strain ZJB12195 with scattered transition, transversion, insertion / deletion mutations.
Table 4 shows the alignments with similarities <97% between
the genome segments of LKHE01000676 of strain 1229
and the transcriptome sequences of GAGW00000000 and
GCQL00000000 of natural C. sinensis and strain L0106 with
scattered transition, transversion, and insertion/deletion mutations
[28,33,53]. Several segments of LKHE01000676 were
transcribed in natural C. sinensis but not in strain L0106, or vice
versa, indicating alternative on-or-off of gene transcription in
natural C. sinensis and strain L0106, or differential post-transcriptional
modifications in response to the natural and unnatural
conditions.
Table 4: Comparison of the genome segment sequences of LKHE01000676 with transcriptome sequences
| Query segment of LKHE01000676 |
Subject transcriptome sequence segment |
% Similarity |
Note |
| Accession # |
Range & direction |
| 1,847→2,330 |
GAGW01005792 |
1→479 |
90.7% (439/484) |
|
| 3,287→3,816 |
GCQL01001639 |
5→513 |
94.9% (504/531) |
|
| 5,386→5,827 |
GAGW01002271 |
1→435 |
97.0% (423/436) |
|
| 6,664→7,561 |
GCQL01010633 |
1→896 |
96.1% (864/899) |
|
| GAGW01002270 |
1←396 |
94.2% (376/399) |
|
| 12,283→12,651 |
GAGW01014001 |
1→370 |
94.4% (353/374) |
|
| GCQL01011474 |
1→285 |
93.8% (271/289) |
|
| 14,111→15,149 |
GCQL01011474 |
283→1,326 |
96.8% (1,014/1,048) |
|
| 17,291→18,242 |
GCQL01000318 |
986→1,326 |
96.5% (329/341) |
|
| GAGW01001243 |
859→1,407 |
96.4% (530/550) |
|
|
518→858 |
96.2% (328/341) |
|
| 19,006→20,474 |
GAGW01001246 |
1←1,414 |
95.0% (1,399/1,472) |
Overlapped with GAGW01001246 |
| GAGW01001244 |
1←260 |
94.2% (245/260) |
| GAGW01001245 |
1←919 |
83.9% (818/975) |
| GCQL01004794 |
1←453 |
89.8% (407/453) |
|
| GCQL01013071 |
8→597 |
87.2% (532/611) |
|
| 20,592→22,337 |
GCQL01006612 |
359→636 |
95.7% (266/278) |
|
|
641→1,272 |
95.6% (605/633) |
|
|
8→359 |
93.3% (334/358) |
|
| GAGW01003631 |
41→318 |
95.7% (266/278) |
|
| 22,376→22,660 |
GCQL01019951 |
1←285 |
96.5% (275/285) |
|
| 28,034→28,569 |
GCQL01010894 |
1→525 |
93.3% (500/536) |
|
| 33,387→34,679 |
GAGW01000758 |
294→1,057 |
93.1% (760/816) |
|
| GAGW01012024 |
2←589 |
96.6 % (568/588) |
Overlapped |
| GAGW01012122 |
1→580 |
91.7% (532/580) |
| 44,841→46,018 |
GCQL01005661 |
1→1,144 |
93.7% (1,117/1,192) |
|
| 50,206→51,036 |
GCQL01003788 |
10→790 |
93.6% (778/831) |
|
| 68,282→69,541 |
GAGW01006524 |
1,751←2,956 |
94.1% (1,187/1,261) |
|
| GCQL01019941 |
1←281 |
82.4% (277/336) |
|
| 92,442→93,636 |
GAGW01003452 |
1→250 |
94.4% (237/251) |
|
| GAGW01003450 |
1←621 |
89.5% (561/627) |
|
| 95,026→95,351 |
GAGW01000444 |
63→320 |
96.9% (250/258) |
Overlapped |
| GAGW01000445 |
1←256 |
96.9% (248/256) |
|
| GAGW01000442 |
1←247 |
96.8% (239/247) |
|
| GAGW01000616 |
1←263 |
95.1% (250/263) |
|
| 107,232→110,427 |
GCQL01005615 |
1→2,902 |
95.8% (2,842/2,967) |
|
| GAGW01009911 |
75→869 |
92.2% (733/795) |
|
| 121,758→123,734 |
GCQL01008379 |
1,124←2,896 |
89.2% (1,764/1,977) |
|
| GAGW01006329 |
1,457→3,231 |
89.2% (1,763/1,977) |
|
| 124,838→125,131 |
GCQL01008379 |
1←288 |
93.6% (277/296) |
|
Genome Sequence LWBQ01000028 of H. sinensis Strain ZJB12195
Segments 80,533→92,360; 104,744→115,873;
118,393→131,981; 134,541→148,041; 162,533→174,118;
185,839→196,118; 226,274→235,333; 235,360→294,143;
249,222→264,107; 276,078→290,663 (and more) of LWBQ01000028 of strain ZJB12195 are 99%-100% homologous
to many segments of ANOV01000226/ANOV01006525,
LKHE01002847, and JAAVMX010000008 of strains Co18, 1229,
and IOZ07, respectively [28,33,53]. Several other segments of
LWBQ01000028, however, share 85.7%-100% similarities with
the genome sequences of strains Co18, 1229, and IOZ07, as
shown in (Figure S4 and Table 5) [26,28,35-36].
Table 5: Comparisons of the genome segment sequences of LWBQ01000028 with other genome sequences
| Query segment of LWBQ01000028 |
Subject genome sequence segment |
% Similarity |
| Accession # |
Range & direction |
| 79,596→82,541 |
ANOV01004731 |
885→3,775 |
96.3% (2,836/2,946) |
| JAAVMX010000003 |
17,109,763←17,112,653 |
93.1% (2,743/2,946) |
| LKHE01000694 |
11,826→14,722 |
92.0% (2,710/2,946) |
| ANOV01001350 |
1←2,895 |
90.8% (2,676/2,946) |
| 84,386→84,802 |
ANOV01002409 |
3→419 |
96.6% (403/417) |
| LKHE01003110 |
106,424→106,839 |
94.5% (393/416) |
| JAAVMX010000003 |
10,251,043→10,251,459 |
94.2% (393/417) |
| 178,302→178,718 |
JAAVMX010000002 |
7,904,858→7,905,270 |
95.2% (397/417) |
| LKHE01001213 |
2,969←3,383 |
94.5% (395/418) |
| ANOV01013978 |
3,090→3,499 |
91.9% (384/418) |
| 179,439→180,080 |
JAAVMX010000009 |
692,799→693,460 |
92.6% (613/662) |
| LKHE01003155 |
2,038←2,699 |
92.6% (613/662) |
| 180,581→180,898 |
LKHE01000683 |
145←462 |
96.5% (307/318) |
| JAAVMX010000007 |
1,986,719→1,987,036 |
96.5% (307/318) |
| 226,274→226,552 |
LKHE01002847 |
40,829→41,085 |
90.0% (252/280) |
| JAAVMX010000008 |
1,836,903←1,837,159 |
| ANOV01006525 |
3,281←3,537 |
| 235,171→235,550 |
LKHE01002847 |
49,706→50,050 |
90.8% (345/380) |
| JAAVMX010000008 |
1,827,939←1,828,283 |
| 290,668→290,946 |
LKHE01002847 |
104,998→105,273 |
94.3% (264/280) |
| JAAVMX010000008 |
1,772,942←1,773,217 |
| ANOV01000226 |
5,870←6,145 |
| 313,425→313,999 |
LKHE01002814 |
9,543→10,117 |
96.7% (556/575) |
| JAAVMX010000006 |
5,835,407←5,835,791 |
91.2% (351/385) |
| ANOV01001029 |
15,460→15,844 |
91.2% (351/385) |
| 315,290→315,616 |
JAAVMX010000008 |
1,748,980←1,749,277 |
89.4% (295/330) |
| LKHE01002814 |
11,291←11,588 |
89.1% (294/330) |
| ANOV01007356 |
1,810→2,154 |
85.7% (299/349) |
| 426,625→427,144 |
JAAVMX010000008 |
1,641,405←1,641,924 |
100% (520/520) |
| ANOV01002726 |
17,746←18,265 |
99.8% (519/520) |
| LKHE01002770 |
149,005←149,524 |
94.2% (490/520) |
| 430,444→430,823 |
JAAVMX010000008 |
1,637,824←1,638,203 |
100% (380/380) |
| LKHE01001032 |
1,568←1,947 |
100% (380/380) |
| ANOV01002726 |
14,167←14,544 |
94.2% (358/380) |
| 507,607→508,491 |
JAAVMX010000008 |
1,563,880←1,564,764 |
99.8% (883/885) |
| LKHE01003593 |
25,479←26,363 |
92.5% (819/885) |
| ANOV01000543 |
11,240←12,124 |
92.1% (815/885) |
| 593,072→593,571 |
LKHE01003291 |
63,024→63,516 |
94.1% (464/493) |
| JAAVMX010000002 |
16,452,980→16,453,478 |
94.1% (464/493) |
| ANOV01010922 |
75→574 |
93.6 % (468/493) |
| 595,906→597,353 |
LKHE01003032 |
4,629→6,071 |
93.9% (1,355/1,443) |
| JAAVMX010000003 |
17,106,470←17,107,917 |
93.5% (1,354/1,448) |
| ANOV01002409 |
4←1,446 |
91.8% (1,329/1,448) |
(Table S4) shows the differentially expressed transcripts in the
transcriptome assemblies of natural C. sinensis and H. sinensis strain L0106. For instance, segment 416,688→417,407
of the genome sequence LWBQ01000028 is 84.6% similar
to GCQL01000179 of strain L0106 with 6 detection gaps
up to 51 bp., but absent from the transcriptome assembly
GAGW00000000 of natural C. sinensis, indicating transcriptional
silencing of the gene in natural C. sinensis and unnatural
transcriptional activation in H. sinensis. Low similarities
(<97%) were found between some of the “paired” transcripts
of natural C. sinensis and strain L0106. Several of these transcripts
from strain L0106 are 2-3-fold in length than the
transcripts from natural C. sinensis, such as GCQL01005624
vs. GAGW01010266, GCQL01006423 vs. GAGW01007466,
GCQL01019079 vs. GAGW01008721, etc., indicating differential
transcription or post-transcriptional modifications in the 2
samples.
Genome Sequence LWBQ01000037 of H. sinensis Strain ZJB12195
Multiple sequences of LWBQ01000037 of strain ZJB12195 are
99%-100% homologous with segments of the genome sequences
ANOV00000000, LKHE00000000, and JAAVMX000000000
of strains Co18, 1229, and IOZ07, respectively [26,28,35-36].
Many short segments of the lengthy sequence LWBQ01000037
are <97% similar to other overlapped genome sequences with
scatted transition, transversion and insertion/deletion mutant
alleles (Figure S5). (Table S5) shows that many segment sequences
of the genome LWBQ01000037 of strain ZJB12195 are
82.0%-96.2% similarities to the numerous segment sequences
of LKHE00000000, JAAVMX000000000, and ANOV00000000 of
strains 1229, IOZ07, and Co18 [26,28,35-36].
(Table S6) compares the genome segment seqeunces of
LWBQ01000037 of strain ZJB12195 with fewer similarities
(80.0%-97.6%) to transcriptome sequences of natural C. sinensis and strain L0106 [26,28,33,35-36,53]. Two genome
segments (14,715→15,238 and 452,296→452,665) of
LWBQ01000037 were transcribed in natural C. sinensis but not
in strain L0106. Some transcripts of natural C. sinensis were 2-3-
fold longer in length than those of strain L0106, corresponding
to the genome segments 6,232→7,009, 39,296→39,287,
and 352,166→352,524 of LWBQ01000037. However, segment
389,632→389,985 of LWBQ01000037 was transcribed >2-fold
longer in strain L0106 than in natural C. sinensis.
The Unassembled Shotgun Genome Sequences
of H. sinensis Strain YN07-8
Five (JM973567, JM973711, JM973713, JM973797, and
JM973816) (1.97%) of the 254 unassembled shotgun genome
sequences (JM973567-JM973820) of strain YN07-8 did not align
to any part of H. sinensis genome and mitogenome sequences
[26,28-29,33,35-36,39,48,53]. Among them, JM973797
and JM973816 are 100% homologous to the transcriptome sequences GAGW01010110 (41→665) and GAGW01003685
(1,636←2,243), respectively, of natural C. sinensis, but absent
from the transcriptome GCQL00000000 of strain L0106 or genome
and transcriptome sequences of other fungal strains registered
in GenBank to date [33,39,53]. However, the upstream
region (1→657) of GAGW01003685 is 97.7% homologous to
the genome segments of JAAVMX010000003, ANOV01000070,
LKHE01001512, and LWBQ01000096 of H. sinensis strains
[26,28,33,35-36,53] IOZ07, Co18, 1229, and ZJB12195, respectively,
whereas GAGW01010110 did not align to any part
of the genome sequences of H. sinensis strains. It seems that
the 5 unassembled genome sequences and the transcript
GAGW01010110, as well as part of GAGW01003685, were
derived from the genomes of fungi co-colonized in natural C.
sinensis other than H. sinensis. The transcript GAGW01003685,
however, might be incorrectly assembled with transcript segments
derived from the heterogeneous fungal genomes in natural C. sinensis.
Six unassembled sequences (2.36%) are ribosomal 28S gene
sequences (Section III.4), 5 sequences (1.97%) are mitogene
sequences (Section III.10), and 163 sequences (64.2%) are
97%-100% homologous to a single copy of each of the assembled
genome sequences of H. sinensis strains [26,28,35-36,39].
Eleven sequences (4.33%) are 98.1%-100% homologous to sequences
of the genome assemblies JAAVMX000000000 and
LKHE00000000 of strains IOZ07 and 1229, respectively, but unmatched
to any part of ANOV00000000 or LWBQ00000000 of
strains Co18 and ZJB12195. Three (1.18%) are 98.6%-99.8% homologous
to a single segment of each of the genome sequences
of JAAVMX000000000, ANOV00000000, and LKHE00000000,
of strains IOZ07, Co18, and 1229, respectively, but JM973722
and JM973755 aligned to double, overlapped LWBQ00000000
sequences of strain ZJB12195.
Thirty unassembled sequences (11.8%) are 57.7%-96.6% similar
to sequences of the genome assemblies ANOV00000000,
LKHE00000000, LWBQ00000000, and JAAVMX000000000 of
strains Co18, 1229, ZJB12195, and IOZ07, respectively, with
scattered transition, transversion, and insertion/deletion point
mutations [26,28,35-36,39]. Eight of the 30 sequences are
57.7%-95.8% similar to >100 overlapped genome sequences;
whereas sequence JM973819 is 94.7%-95.2% similar to 38
overlapped genome sequences.
Thirty one unassembled sequences (12.2%) are ≥ 97% homologous
to some of the assembled genome sequences but less
similar (<97%) to other sequences with scattered transition,
transversion, and insertion/deletion mutant alleles [26,28,35-36,39]. Six of them aligned to >100 overlapped sequences of
the assembled genomes with various similarities of 79.9%-
99.8%.
Transcription of the Unassembled Genome Sequences
of H. sinensis Strain YN07-8
Sixteen (6.29%) of the 254 unassembled genome sequences did not align to any transcriptome sequences of natural C. sinensis or H. sinensis strain L0106, indicating that these genome
components were non-transcriptable or transcriptionally silent.
Sixty-three sequences (24.8%) were expressed differentially
in strain L0106 but not in natural C. sinensis; whereas 6
other sequences (2.36%) expressed in natural C. sinensis but
not in strain L0106 and might be derived from fungi colonized
in natural C. sinensis other than H. sinensis.
Among those unassembled genome sequences that are highly
homologous to the assembled genome sequences of H. sinensis strains, 70 sequences (27.6%) were transcribed to a single
transcript copy of strain L0106 and natural C. sinensis, whereas
46 (18.1%) transcribed in either strain L0106 or natural C. sinensis but not both [33,39,53]. Forty-seven (18.5%) of the unassembled
sequences were transcribed with multiple overlapped
transcripts in strain L0106 but 27 (10.6%) transcribed with multiple
overlapped transcripts in natural C. sinensis. For instance,
JM973748 aligned to 94 overlapped transcripts of natural C.
sinensis and 6 overlapped transcripts of strain L0106.
The OSRC14 Marker Genes of H. sinensis Strains
Sequence similarities of 95.2%-100% were found among the
PCR-amplified OSRC14 gene sequences of 42 H. sinensis strains
(Table 6) [40]. Among them, JQ277392, JQ325408, JQ325431,
JQ325442-JQ325443 and KM197544 contain numerous mismatched
alleles (Figure S6). JQ325431 of strain XZ05-8 is
92.2%-95.2% similar to the OSRC14 sequences of 35 H. sinensis strains.
Table 6: Comparisons of the OSRC14 gene sequence JQ325484 (the Query) of strain YN09-140 with other OSRC14 gene sequences (the Subject)
| Accession # |
H. sinensis strain |
% Similarity vs. JQ325484 (strain YN09-140) |
| JQ277386 |
QH07-197 |
100% (577/577) |
| JQ277389 |
QH09-93 |
100% (577/577) |
| JQ277390 |
XZ07-176 |
100% (577/577) |
| JQ325373 |
GS09-121 |
100% (577/577) |
| JQ325377 |
GS09-225 |
100% (577/577) |
| JQ325402 |
QH09-210 |
100% (577/577) |
| JQ325422 |
SC09-190 |
100% (577/577) |
| JQ325429 |
XZ05-6 |
100% (577/577) |
| JQ325438 |
XZ07-133 |
100% (577/577) |
| JQ325451 |
XZ08-59 |
100% (577/577) |
| JQ325458 |
XZ09-48 |
100% (577/577) |
| JQ325462 |
XZ09-95 |
100% (577/577) |
| JQ325473 |
YN09-6 |
100% (577/577) |
| JQ325474 |
YN09-22 |
100% (577/577) |
| JQ325476 |
YN09-61 |
100% (577/577) |
| JQ325477 |
YN09-64 |
100% (577/577) |
| JQ325481 |
GS09-311 |
100% (577/577) |
| JQ325482 |
YN09-96 |
100% (577/577) |
| JQ325485 |
ID10-1 |
100% (577/577) |
| JQ325486 |
NP10-1 |
100% (577/577) |
| JQ325487 |
NP10-2 |
100% (577/577) |
| JQ325397 |
QH09-151 |
99.8% (576/577) |
| JQ325398 |
QH09-164 |
99.8% (576/577) |
| JQ325461 |
XZ09-80 |
99.8% (576/577) |
| JQ325472 |
YN09-3 |
99.8% (576/577) |
| JQ325475 |
YN09-51 |
99.7% (575/577) |
| JQ277391 |
SC09-37 |
98.8% (570/577) |
| JQ325409 |
SC09-47 |
98.8% (570/577) |
| JQ325455 |
XZ09-15 |
98.8% (570/577) |
| JQ325464 |
XZ09-106 |
98.8% (570/577) |
| JQ325478 |
YN09-72 |
98.8% (570/577) |
| JQ325479 |
YN09-81 |
98.8% (570/577) |
| JQ325480 |
YN09-85 |
98.8% (570/577) |
| JQ325483 |
YN09-101 |
98.8% (570/577) |
| JQ325481 |
YN09-89 |
98.6% (569/577) |
| JQ325408 |
SC09-36 |
96.5% (557/577) |
| JQ277392 |
XZ06-124 |
95.4% (561/588) |
| JQ325443 |
XZ07-H2 |
95.4% (561/588) |
| JQ325431 |
XZ05-8 |
95.2% (560/588) |
| JQ325442 |
XZ07-H1 |
95.2% (560/588) |
| KM197544 |
XZ12-16 |
95.2% (560/588) |
JQ325484 of strain YN09-140 is 100% homologous to the assembled
genome segments 34,066→34,642 of LKHE01001606,
649,477→650,053 of JAAVMX010000011, and 6,294→6,870
of ANOV01000797 of strains 1229, IOZ07, and Co18, and
52,641←52,065 and 245,347→245,923 of LWBQ01000349 and
LWBQ01000064 of strain ZJB12195, respectively [26,28,35-36,40].
No matches were found between OSRC14 sequences and the
GCQL00000000 transcriptome assembly of strain L0106, indicating
transcriptional silencing of the OSRC14 gene in H. sinensis [33,40]. Segments 1→67 and 364→577 of OSRC14 sequence
JQ325484 of strain YN09-140 is 100% homologous to
segments 451←517 and 78←291 of the GAGW01003073 transcriptome
assembly of natural C. sinensis but another segment
(125→295) of JQ325484 is 97.1% similar to segment 282←452
of GAGW01003073 with scattered transition and transversion
point mutations [40,53]. Considering the integrity of PCR-amplified
sequence JQ325484, the 3 segments of GAGW01003073
might have been assembled with heterogeneous shotgun transcripts
derived from the genomes of independent fungi in natural C. sinensis. If OSRC14 gene is transcriptionally silent in H.
sinensis, the 3 assembled GAGW01003073 transcripts might be
derived from the OSRC14 genes of independent fungi in natural C. sinensis other than H. sinensis.
The OSRC19 Marker Genes of H. sinensis Strains
The unassembled shotgun genome segment JM973741 (the
OSRC19 marker gene) of H. sinensis strain YN07-8 is 99.8%
homologous to the PCR-amplified OSRC19 gene sequences
JQ277405 and JQ277406 of strains XZ06-124 and SC09-37 but
only 94.5% similar to the OSRC19 sequences JQ277407 and
JQ277408 of strains QH09-93 and XZ07-176, respectively, with
scattered transition and transversion point mutations (Figure
S7) [39-41].
JM973741 of strain YN07-8 is 94.5%-98.6% homologous to
segments 2,470→3,201 of ANOV01007159; 55,477←56,202 of LKHE01000676; 54837←55562 of JAAVMX010000004 of
strains Co18, 1229, and IOZ07, respectively, and non-overlapped
segments 140,812→141,235 and 141,367→141,660 of
LWBQ01000084 of strain ZJB12195, with scattered transition and transversion, and insertion/deletion mutant alleles (Figure
3) [26,28,35-36,39-40]. A 131-nt (141,236→141,366) DNA segment
deletion was found between the 2 aforementioned segments
of LWBQ01000084.

Figure 3: Alignment of the genome assembly segments LKHE01000676 with other genomic sequences.
JM973741 did not align to any sequences of the transcriptome
assemblies GAGW00000000 of natural C. sinensis and
GCQL00000000 of strain L0106 and may represent a non-transcribed
intron sequence or a silent gene.
Cross-analysis revealed that the lengthy segment 827→117,239
of LKHE01000676 of strain 1229 is 99.8% homologous to
segment 191→116,599 of JAAVMX010000004 of strain
IOZ07, and 98.4%-98.9% similar to much shorter segments
of LWBQ01000084 (104,348←125,987) of strain ZJB12195
and ANOV01007159 (1←7,193) of strain Co18 [26,28,35-36]. Segment 827→1,348 of LKHE01000676 is 74.2%-91.9%
similar to segments 140,304→140,677 of LWBQ01000135,
1,608←2,119 of ANOV01006005, 191→718 and 483→909
of JAAVMX010000004, and 4,008,152←4,008,663 of
JAAVMX010000001 of strains ZJB12195, Co18, and IOZ07, respectively.
Segment 191→909 of JAAVMX010000004 is 97%
similar to segment 342→1068 of LKHE01000676 of strain 1229
but 82% similar to segments 1,252←2,124 of ANOV01006005
and 140,304→141,471 of LWBQ01000135 of strains Co18 and
ZJB12195, respectively, with scattered transition, transversion,
and inversion/deletion point mutations.
The OSRC27 Marker Genes of H. sinensis Strains
Sequence similarities of 93.8%-100% were found among the
PCR-amplified OSRC27 gene sequences of 38 H. sinensis strains
(Table S7 and Figure S8) [40,42]. JQ325705 of strain YN09-6 is
92.6%-94.2% similar to other OSRC27 sequences of 32 H. sinensis strains with scattered transition, transversion, and insertion/
deletion point mutations.
JQ325719 of strain NP10-2 is 100% homologous to the assembled
genome sequences, ANOV01003376, LKHE01000526,
LWBQ01000003, and JAAVMX010000005 of H. sinensis strains
Co18, 1229, ZJB12195, and IOZ07, respectively [26,28,35-36,40,42]. JQ325705 of strain YN09-6 is only 93.8% similar to
the OSRC27 genome sequences of the H. sinensis strains.
The OSRC27 gene was not transcribed in natural C. sinensis.
Segments 1→135 and 240→574 of JQ325719 of strain NP10-
2 are 99.7%-100% homologous to segments 1,651←1,985 and 1,983←2,217 of transcriptome sequence GCQL01013271 of
strain L0106 with a 104-bp. segment deletion, corresponding to
the genome segment 136→239 of JQ325719 [33,42]. Segments
1,983←2,217 of GCQL01013271 is completely overlapped with
20←154 of GCQL01015312 of strain L0106. JQ325705 of strain
YN09-6 is 97.3%-98.5% similar to the 2 transcript segments
of GCQL01013271 with scattered transition and transversion
point mutations (Figure S9) [33,40,42].
The OSRC32 Marker Genes of H. sinensis Strain
The unassembled genome sequence JM973601 (the OSRC32
marker gene) of strain YN07-8 is 88.6%-100% similar to other
PCR-amplified OSRC32 gene sequences of 35 H. sinensis strains (Table 7 and Figure S10), with scattered transition and
transversion mutant alleles and a DNA segment insertion/deletion
[39-41]. JM973601 is 91.8-93.1% similar to the assembled
genome segments 378,579←379,528 of LWBQ01000050,
32,632←33,579 of LKHE01001701, 2,495←3,442 of
ANOV01000094, and 452,225→453,180 of JAAVMX010000004
of strains 1229, ZJB12195, Co18, and IOZ07, respectively, with
scattered transition, transversion, and insertion/deletion point
mutations (Figure 4) [26,28,35-36,39].
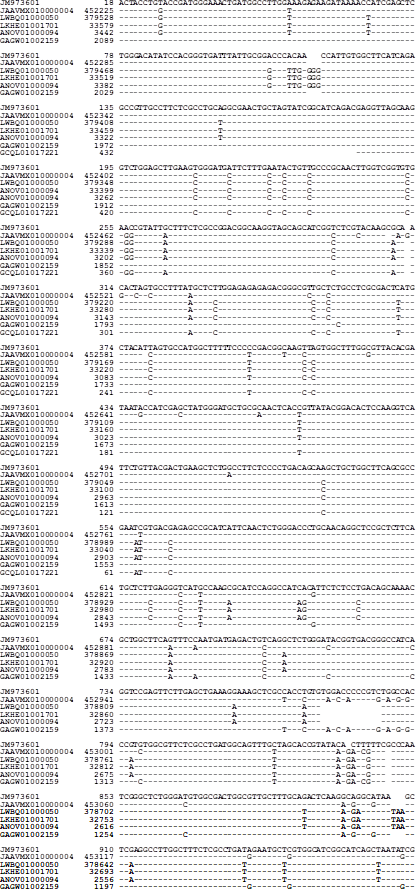
Figure 4: Alignment of the genome assembly segments LKHE01000676 with other genomic sequences.
(Table 7)Cross-analyses revealed that ANOV01000094 of strain
Co18 is 95.7%-99.6% homologous to the genome segments
of LWBQ01000050 (376,024→384,921; 385,007→386,603;
386,612→392,750; 392,856→394,301; 394,358→399,049),
LKHE01001701 (30,138→52,637), and JAAVMX010000004
(433,098←454,779; 454,788←455,675) of strains ZJB12195,
1229, and IOZ07, respectively [26,28,35-36]. Some of the genome
sequences contain scattered transition, transversion,
and insertion/deletion mutant alleles.
(Figure 4) also shows that JM973601 of strain YN07-8 is 94.2%-
96.5% similar to the transcriptome sequences GCQL01017221
(1←432) of strain L0106 and GAGW01002159 (1,134←2,106)
of natural C. sinensis with scattered transition, transversion,
and insertion/deletion mutations [33,39,53]. The segment
1→432 of GCQL01017221 is only 94.0% similar to the segment
1,493→1,924 of GAGW01002159.
Segment 1→2,089 of GAGW01002159 of natural C. sinensis is
98.3% homologous to the genome sequence 452,225←454,313
of JAAVMX010000004 of strain IOZ07. However, a shorter segment
452,390←452,821 of JAAVMX010000004 is only 94.9%
similar to the transcriptome sequence GCQL01017221 of
strain L0106, whereas GCQL01017221 is 100% homologous
to other genome sequences 2,843→3,274 of ANOV01000094,
378,929→379,360 of LWBQ01000050, and 32,980→33,411
of LKHE01001701 of strains Co18, ZJB12195, and 1229, respectively
[26,28,33,35-36]. In contrast, GAGW01002159
(419→2,949) of natural C. sinensis is only 87.4% similar to the
genome segment sequences: ANOV01000094 (1,778→4,302),
LWBQ01000050 (377,864→380,388), and LKHE01001701
(31,915→34,439) of strains Co18, ZJB12195, and 1229, respectively,
with scattered transition, transversion, and insertion/
deletion point mutations. It is questionable whether the full or
segmented transcript GAGW01002159 were derived from the
genome (s) of one or more of the fungi co-colonized in natural C. sinensis other than H. sinensis, shows segment insertions/
deletions in multiples of 3 to indicate open reading frames of
proteins.
The OSRC11, OSRC23, and OSRC31 Marker
Gene Sequences of H. sinensis Strains
Sequences JQ277384 (OSRC11 gene) of strain XZ06-124 and
JQ277444 (OSRC31 gene) of strain XZ07-H2 are 99.5%-99.8% homologous
to the assembled genome sequences ANOV01006466
and LKHE01002656 of strains Co18 and 1229 but absent from
the genomes LWBQ00000000 and JAAVMX000000000 of
strains ZJB12195 and IOZ07, respectively [26,28,35-36,39]. The
OSRC23 gene sequence JQ277420 of strain XZ07-H2 is 99.2%
homologues to LKHE01002410 of strain 1229 but absent from
the genome assemblies JAAVMX000000000, LWBQ00000000,
and ANOV00000000 of strains IOZ07, ZJB12195, and Co18, respectively.
Sequences of JQ277384 (OSRC11 gene) and JQ277420 (OSRC23
gene) are absent from the transcriptome assembly of strain
L0106 but the genes were transcribed in natural C. sinensis to
35 and 352 overlapped GAGW00000000 transcripts, respectively,
with similarities of 82.7%-100% [35,39,53]. The OSRC31
gene sequence JQ277444 is 99.3%-99.5% homologous to the
transcriptome sequences of natural C. sinensis and strain L0106.
The mRNA Sequence KP090937 of H. sinensis Strain L0106
Both the hexokinase-like mRNA sequence KP090937 and assembled
transcriptome sequence GCQL01012008 (1←1,137)
were from H. sinensis strain L0106 and are 100% identical
(Figure 5) [33]. Two segments (1→326 and 328→1,137) of
KP090937 are 99.7%-100% homologous to the non-overlapped
transcripts 1←326 of GAGW01010481 and 1→810 of
GAGW01005022, respectively, of natural C. sinensis.
Segments 1→104 and 130→541 of the mRNA sequence
KP090937 of strain L0106 are 100% identical to the genome
segments of LKHE01001829, LWBQ01000186/LWBQ01000017,
JAAVMX010000002, and ANOV01015216 of strains 1229,
ZJB12195, IOZ07, and Co18, respectively [26,28,33,35-36].
Segment 541→1,137 of KP090937, however, is 87.8%-90.4%
similar to segments of LKHE01001829, LWBQ01000186/
LWBQ01000017, JAAVMX010000002, and ANOV01003538
of strains 1229, ZJB12195, IOZ07, and Co18, respectively,
with 9 deletions in the mRNA sequence KP090937, likely indicating
alternative splicing [26,28,33,35-36,53]. It seems that
ANOV01015216 and ANOV01003538 of strain Co18 might be
incorrectly assembled.
Cross-analysis revealed that the lengthy genome sequences
LKHE01001829 of strain 1229 is 97.6%-100% homologous
to the genome segment sequences of JAAVMX010000002,
LWBQ01000017, and ANOV01000772 of strains IOZ07,
ZJB12195, and Co18, respectively [26,28,35-36]. However, segment
93,470→94,948 of the lengthy sequence LKHE01001829
is 98.1%-98.4% homologous to segments 1→1,214 of
ANOV01015216 of strain Co18 and 18,187,481→18,188,936
of JAAVMX010000002 of strain IOZ07, but only 90.4%-94.7%
similar to segments 174,857→176,321 of LWBQ01000186 and
186,085←187,643 of LWBQ01000017 of strain ZJB12195 with
several insertions/deletions (mono-, bi-, or poly-bases), and
transition and transversion point mutations.
The mRNA Sequence KP090945 of H. sinensis Strain L0106
The ADP-ribose pyrophosphatase-like mRNA sequence
KP090945 of strain L0106 is 100% homologous to the assembled
transcriptome sequences GCQL01000385 and GCQL01014864
of strain L0106 but absent from the transcriptome assembly
GAGW00000000 of natural C. sinensis [33,53], indicating transcriptional
silencing of the ADP-ribose pyrophosphatase gene
in natural C. sinensis but anti-natural activation of the gene in
strain L0106.
KP090945 is 100% homologous to genome sequences
LKHE01001747 (109,170→109,895), JAAVMX010000012
(65,952→66,677), and ANOV01003103 (1←385, 435←620)
of strains 1229, IOZ07, and Co18, respectively, but only 94.1%
similar to segment 122,131→122,896 of LWBQ01000044 of
strain ZJB12195 with 2 large segments of DNA insertions/deletions
[26,28,33,35-36].
Cross-analysis revealed that the lengthy genome segment of
LWBQ01000044 of strain ZJB12195 is 99.3%-99.6% homologous to segments of JAAVMX010000001/JAAVMX010000012,
LKHE01000716/LKHE01001747, and ANOV01000098/
ANOV01005573 of strains IOZ07, 1229, and Co18, respectively
[26,28,35-36]. However, segment 120,984→122,545
of LWBQ01000044 is 93.5%-95.2% similar to segments
108040→109544 of LKHE01001747, 64,802→66,326 of
JAAVMX010000012, and 1←1,151 of ANOV01003103 of strains
1229, IOZ07, and Co18, respectively, with several DNA insertions/
deletions and some transition and transversion point
mutations (Figure S11) [26,28,35-36].
The mRNA Sequence KP090946 of H. sinensis Strain L0106
The ribose-phosphate pyrophosphokinase-like mRNA sequence
KP090946 of strain L0106 is 99.8%-100% homologous
to the transcriptome assemblies GCQL01011182 and
GAGW01003914 of strain L0106 and natural C. sinensis, respectively
(Figure S12) [33,53].
KP090946 is 96.4%-96.5% similar to the genome segments
4,761→6,137 of LKHE01001673, 5,597,720←5,599,096 of
JAAVMX010000006, and 20,514←21,890 of ANOV01001719
of strains 1229, IOZ07, and Co18, respectively, with a 48-nt.
deletion occurred in the sequence of KP090946, as well as
in GCQL01011182 and GAGW01003914 (Figure S12), indicating
alternative splicing during transcription [26,28,33,35-36,53]. KP090946 is only 89.6% similar to LWBQ01000045
(385,652→386,959) of strain ZJB12195 with 4 segment insertions/
deletions and scattered transversion and transition point
mutations [33,35].
Cross-analysis revealed that segments 1→10,761 and
10,706→23,317 of ANOV01001719 of strain Co18 is 99.7%-
99.8% homologous to segments 15,855←26,615 and
23,334←15,917 of LKHE01001673 of strain 1229 and segments
5,577,280→5,588,040 and 5,587,978→5,600,523
of JAAVMX010000006 of strain IOZ07 [26,28,36]. Segment
6,493→10,761 of ANOV01001719 is 99.1% homologous to segment
397,504←401,738 of LWBQ01000045 of strain ZJB12195,
but many other ANOV01001719 segments are 93.2%-96.6%
similar to the segments of LWBQ01000045 [26,35].
The mRNA Sequence KP090949 of H. sinensis Strain L0106
The amidophosphoribosyl transferase-like mRNA sequence
KP090949 is 100% homologous to the 2←952 of GCQL01009009
transcriptome assembly of strain L0106 [53]. In contrast to the
full-length transcript in strain L0106, the gene was partially
transcribed in natural C. sinensis to a segment 176→514 of
GAGW01013335 with a 613-nt. deletion corresponding to segment
339→951 of KP090949, indicating alternative transcription
or posttranscriptional change in response to the natural
and unnatural conditions [33,53].
KP090949 is 93% similar to genome sequences
424,486←425,462 of LWBQ01000048, 15,195,213←15,196,189
of JAAVMX010000003, 7,439←8,415 of LKHE01001105,
9,085←10,061 of ANOV01001694 of strains ZJB12195, IOZ07,
1229, and Co18, respectively [26,28,33,35-36]. The most variable
segment 403→448 of KP090937 consists of multiple transition, transversion, and DNA segment deletions (mono-, bi- or
poly-bases) in the KP090946 mRNA sequence, some of the
deletions are not in multiples of 3 (Figure S13). Cross-analysis
revealed high homologies (98.8%-99.7%) among the 4 assembled
genome sequences LKHE01001105, LWBQ01000048,
JAAVMX010000003, and ANOV01001694 of H. sinensis strains
1229, ZJB12195, IOZ07, and Co18, respectively [26,28,35-36].
The mRNA Sequence KP090959 of H. sinensis Strain L0106
The 5’-nucleotidase-like mRNA sequence KP090959 of strain
L0106 is 100% and 99.8% homologous to transcriptome sequences
GCQL01011621 of strain L0106 and GAGW01006965
of natural C. sinensis (Figure S14) [33,53].
Two segments (1←333 and 333←1,971) of KP090959
of strain L0106 are 86.9%-96.1% similar to segments
9,371,542←9,373,237 and 9,373,304←9,373,686 of
JAAVMX010000005; 88,957←90,652 and 90,719←91,101
of LKHE01002574; and 15,089←16,784 and 16,851←17,233
of ANOV01001374 of strains IOZ07, 1229, and Co18, respectively,
with scattered transition, transversion, and insertion/
deletion point mutations (Figure S14) [26,28,33,35-36].
However, KP090959 is 86.9%-100% similar to 5 segments
of LWBQ01000131 of strain ZJB12195: 206,165→206,547;
206,614→207,046; 207,045→207,352; 207,353→207,564;
and 207,574→208,657 [33,35]. As the result of transcription
splicing process, a 67-nt. deletion occurred between the 2 segment
sequences of KP090959, locating between the genome
segment sequences of JAAVMX010000005, LKHE01002574,
and ANOV01001374 and between nucleotides 206,547 and
206,614 of LWBQ01000131.
Cross-analysis revealed that the genome sequence of
ANOV01001374 of strain Co18 is 99.2%-99.9% homologous
to the genome sequences of JAAVMX010000005 and
LKHE01002574 of strains IOZ07 and 1229, respectively
[26,28]. However, Table S8 shows many segment sequences
of ANOV01001374 with 90.9%-96.6% similarities to many segments
of LWBQ01000131 of strain ZJB12195, in addition to 70
other high-similarity LWBQ01000131 segments (>97%) with
large segment insertions/deletions and scattered transversion
and transition point mutations.
The mRNA Sequence KP090961 of H. sinensis Strain L0106
The purine nucleosidase-like mRNA sequence KP090961 is
97.9% and 100% homologous to the overlapped transcriptome
sequences GCQL01017603 and GCQL01014666 of strain L0106
but absent from the GAGW00000000 transcriptome assembly
of natural C. sinensis [33,53], indicating transcriptional silencing
of the gene in natural C. sinensis and anti-natural transcriptional
activation in strain L0106.
KP090961 is 98.2%-98.9% homologous to segments
64,831←65,280 of LKHE01002043 and 15,187,215←15,187,928
of JAAVMX010000003 of strains 1229 and IOZ07, only 93.0%
similar to segment 416,051←416,805 of LWBQ01000048 of
strain ZJB12195, but absent from the ANOV00000000 genome
assembly of strain Co18 [26,28,33,35-36]. Comparing to the 3 genome sequences, there is an 8-nt. deletion between nucleotides
539 and 540 of the mRNA sequence KP090961. There
is a 41-nt. insertion in the LWBQ01000048 genome sequence,
locating between nucleotides 378 and 379 of the KP090961 sequence.
Upstream of the inserted segment in LWBQ01000048,
3 genome segment sequences of strains share 100% sequence
homology.
Cross-analysis revealed that LKHE01002043 of strain 1229
is 97.8%-99.4% homologous with multiple overlapped segments
of ANOV01005037/ANOV01006908/ANOV01012844,
LWBQ01000048, and JAAVMX010000003 [26,28,35-36].
LKHE01002043 is 74.6%-96.4% similar to numerous other segments
of ANOV01000934/ANOV01003645/ANOV01005037/
A N O V 0 1 0 0 5 0 3 8 /A N O V 0 1 0 0 5 5 2 1 /A N O V 0 1 0 0 6 9 0 8 /
ANOV01007855/ANOV01012844, LWBQ01000048/
LWBQ01000085, and JAAVMX010000003 of strains Co18,
ZJB12195, and IOZ07, respectively, with multiple insertions/
deletions and scattered transversion and transition point mutations,
the majority of which form 2 overlapped groups of the
transcripts (Table S9).
Discussion
Transcription of H. sinensis Genes
Liu et al [33]. reported the dynamic transcriptional alteration
in the mycelia of H. sinensis strain L0106 following 3, 6, and
9 days of liquid fermentation without an insect host, featuring
with nonlinear reductions in the total number of transcriptomic
unigenes (25,511→25,214→16,245), nonlinear increases
(681-nt.→682-nt.→994-nt.) in the average unigene length,
and reductions (58.2%→57.9%→57.0%) in the GC content.
Such dynamic alteration indicates switching on or off of multiple
genes in response to continuous in vitro fermentation. Our
cross-analysis of the transcriptome sequences of H. sinensis strain L0106 and natural C. sinensis confirmed differential transcriptional
activation and deactivation and posttranscriptional
modifications of many genes.
A) Many genes were differentially transcribed partially or
in full length in natural C. sinensis but not in H. sinensis strain
L0106 (Sections III.5-III.6, III.10, III.12, III.14-III.17, III.21), or vice
versa (Sections III.5, III.11-III.13, III.16, III.18-III.19, III.23, III.27).
Some transcripts might be derived from the colonized fungi in
natural C. sinensis, other than H. sinensis. Some naturally silent
genes were anti-naturally activated in H. sinensis strain L0106
in respond to the 3-9 days of liquid fermentation without an
insect host. Many other genes were naturally transcripted but
silent unnaturally in strain L0106 [33];
B) Some of the segment sequences of the transcriptome
GAGW00000000 and GCQL00000000 are highly variable
[33,53]. Many transcripts from strain L0106 are 2-3-fold longer
in legth than those from natural C. sinensis, or vice versa, indicating
differential transcription or post-transcriptional modification;
C) Some of the insertion/deletion mutations in the genome
sequences are present in multiples of 3 (Figures 3-4, S2,
S9-S10), which are representative of the protein codons in the
open reading frames for derivation of the amino acid sequences. However, other mutations do not show this pattern (Figures
2, 5, S1, S3-S6, S8, S11-S14), indicating possibilities of translational
disruption of the coding sequences, causing irreversible
arrest of protein translation.
The transcriptome assembly GAGW00000000 was derived
from a natural C. sinensis specimen (unknown maturation
status), which was purchased from the market of Kangding of
Sichuan Province [53]. Liu et al [63]. constructed 2 cDNA libraries
from total mRNA of the stroma and caterpillar body of a C.
sinensis specimen (unknown maturation status) also collected
from Kangding, Sichuan and found apparent differences between
the cDNA libraries. Xia et al [62]. reported a transcriptome
project of the fully mature natural C. sinensis specimens
that were collected from Deqin of Yunnan province. Zhong et al
[60]. reported a transcriptome project of natural C. sinensis “in
the early teleomorph stage without ascus forming” that was
collected from Yushu of Qinghai province. Li et al [64]. reported
another transcriptome project of artificial C. sinensis specimens
and described dynamic transcriptional alterations of many C.
sinensis genes in the different development phases. However,
Liu et al. [63], Xia et al. [62], Zhong et al. [60], and Li et al
[64]. did not upload their assembled transcriptome sequences
in GenBank. Some of them uploaded the assembled cDNA sequences
to www.plantkingdomgdb.com/Ophiocordyceps_sinensis/
but the database is unaccessable for our cross-analysis.
Dong et al. [65] reported significant differences and dynamic
alterations in proteomic polymorphisms between the stroma
and caterpillar body of C. sinensis during maturation, indicating
the diversified and dynamically altered transcriptome profiles
in the C. sinensis stroma and caterpillar body, reflecting the
occurrence of transcriptional, posttranscriptional, and translational
alterations and posttranslational modifications in the
compartments of C. sinensis during maturation.
The Mating-Type Genes
Bushley et al. [24] reported detection of the MAT1-1-1, MAT1-
1-2, and MAT1-1-3 genes of MAT1-1 idiomorph and the MAT1-
2-1 gene of MAT1-2 idiomorph in strain CS68-2-1229 and hypothesized
pseudohomothallism for H. sinensis (Genotype #1
of O. sinensis). Hu et al. [26] reported detection of the MAT1-
1-1 and MAT1-2-1 genes in the genome of strain Co18 and
hypothesized homothallism for H. sinensis. Zhang et al. [66],
however, reported detection of MAT1-2-1 gene, but not genes
of MAT1-1 idiomorph, in H. sinensis strains CS2 and SCK05-4-3.
We summarized in Table 3 the differential occurrence of the
mating genes in H. sinensis strains. Because of the differential
occurrence of homothallic and heterothallic reproduction
strategies, Zhang and Zhang [47] proposed a facultative hybridization
hypothesis for H. sinensis. We found that the mating
genes of MAT1-1 idiomorph were absent from the genome
assembly LWBQ00000000 of strain ZJB12195 and the MAT1-
2-1 gene was absent from JAAVMX000000000 of strain IOZ07.
Such inconsistent occurrence of the mating genes of the MAT1-
1 and MAT1-2 idiomorphs in the genomes of H. sinensis strains
fails to support genetic capability of self-fertility but supports
heterothallic reproduction [67].
In contrast to the occurrence of mating genes in the genomes of H. sinensis strains, the presence of the mating genes of MAT1-1
and MAT1-2 idiomorphs has been reported in several studies of natural C. sinensis that contains multiple co-colonized fungi.
The mating genes were detected (1) in the whole-genome of
fully matured natural C. sinensis [62]; (2) in the early-developed
stroma and caterpillar body of natural C. sinensis with very low
read count values and in 31 other C. sinensis specimens [60],
and (3) in artificial C. sinensis of different development phases
[64]. Although the hyphae of artificial C. sinensis were included
as a development stage of artificial C. sinensis, no methodology
for fungal purification was described [64], indicating that
the hyphae were wild-type fungi as previously reported by
Zhang et al. [16] and Xia et al. [34]. Because of the co-extence
of multiple colonized fungi in natural and artificial C. sinensis,
the results from these studies may not be specifically used to
accurately estimate the genetic capability of the self-fertility of H. sinensis (Genotype #1 of O. sinensis).
Technically self-fertility may come true when the mating genes
of both MAT1-1 and MAT1-2 idiomorphs are transcribed and
translated, and both mating proteins are fully activated within
a single fungal cell. However, our analysis herein demonstrated
that the MAT1-1-1 gene transcript was absent, but MAT1-2-1
gene transcript precent, in the GCQL00000000 transcriptome
assembly of strain L0106 [33,53]. Notably, Zhang and Zhang
[47] observed the 4.9%-6.1% intraspecific variations of MAT1-
1-1 and MAT1-2-1 genes in various H. sinensis strains, which
probably cause the coding sequence disturbance of the genes
and translation arrest and reproductive impotence of H. sinensis.
These findings are inconsistent with the homothallic/
pseudohomothallic mating hypotheses that were underpinned
solely by the genome data but indicate functional heterothallic
behaviors.
Studies have reported variable transcription of the mating
genes in natural and artificial C. sinensis that contains multiple
co-colonized fungi.
1. MAT1-1-1 gene was expressed in all 5 specimens of natural C. sinensis (maturing stage of development), but MAT1-2-1 gene expressed in only 2 of 5 C. sinensis specimens and
MAT1-1-3 gene expressed in only one specimen [68].
2. MAT1-1-3 and MAT1-2-1 genes, but not MAT1-1-1 and
MAT1-2-1 genes, were expressed with very low read count
values in the early-developed stroma and caterpillar body
of natural C. sinensis [60].
3. The 4 mating genes were incoordinately expressed in all
development phases of artificial C. sinensis [64]. They
were expressed in primordium differentiation and mature
fruiting body of artificial C. sinensis, with expression highest
in the fertile part and lowest in the caterpillar body of
mature artificial C. sinensis.
4. Zhao et al. [69] reported nearly no expressions of the
MAT1-1-1 (transcripts per million reads, TPM of 0-2.27)
and MAT1-2-1 (TPM of 0-1.74) genes and concluded that
these genes may not play roles in the fruiting body initiation
stage of C. sinensis.
The differential expression of the mating genes of both MAT1-
1 and MAT1-2 idiomorphs in natural and artificial C. sinensis is then insufficient to prove that the transcripts were from a
single fungal cell of H. sinensis, nor to support the homothallic/
pseudohomothallic mating hypotheses for H. sinensis.
To resemble the functional heterothallic reproduction, H. sinensis (the postulated anamorph and Genotype #1 of o.sinensis)
needs an opposite mating partner capable of expression of the
mating genes of the MAT1-1 and MAT1-2 idiomorphs, meaning
that the telemorphic O. sinensis may have more than one
anamorphic fungus. The candidates of mating partner are the
AT-biased O. sinensis genotype fungi or other fungal species
within one of heterokaryotic cells [24], triggering revisit of the
following academic puzzles observed previously:
A) Hu et al. [26] concluded that the fruiting body and
ascospore productions have been consistently failed after inoculating
ghost moth larvae of Hepialidae family with pure H.
sinensis. Many such inoculation strategies could induce death
and mummification of larvae, but no stromal formation;
B) Wei et al. [70] reported a species contradiction between
the inoculant H. sinensis (GC-biased Genotype #1 of O.
sinensis) and the sole teleomorph of AT-biased Genotype #4 of O. sinensis in the fruiting body of artificial C. sinensis;
C) Co-occurrence of the multiple AT-biased genotypes of O. sinensis and GC-biased H. sinensis (Genotype #1 of O. sinensis)
in the stroma, caterpillar body, ascocarps, and ascospores
of natural C. sinensis [6,8-11,17-19,21-22]. The sequences of
the AT-biased genotypes do not reside in the genome of GC-biased H. sinensis but belong to the genomes of independent
fungi;
D) Mao et al. [32] observed “H”-type fusions of hyphae
during germination, containing AT-biased Genotype #4 or #5
of O. sinensis fungi without GC-biased H. sinensis in natural C.
sinensis specimens collected from different production areas;
E) The biomasses of the AT and GC-biased genotypes of O. sinensis underwent dynamic alterations in an asynchronous
manner in the caterpillar body and stroma of C. sinensis during
maturation, whereas the AT-biased genotypes of O. sinensis always
predominate in the C. sinensis stroma [6,17-19];
F) Two Genotypes #13-14 of O. sinensis were found in
the multicellular heterkaryotic ascospores of natural C. sinensis with mono-, bi-, and tri-nuclear structure. They feature with
reciprocal substitutions of large DNA segments between 2 parental
fungi, Group-A H. sinensis and a Group-E fungus [6,9-11,19,24];
G) Barseghyan et al. [20] reported that Tolypocladium
sinensis and H. sinensis were the dual anamorphs of O. sinensis;
H) The close relationship and tight association of Paecilomyces
hepiali with H. sinensis in the stroma, caterpillar body,
ascocarps, and ascospores of natural C. sinensis and in the intestines
of healthy larvae of Hepialidae family [6,8-9,17-19,30].
Marker Genes Used in Multigene Analysis
Multigene examination strategy has been used in phylogenetic
studies of O. sinensis and the sequences of many DNA loci, nrSSU,
nrLSU, MAT1-1-1, MAT 1-2-1, tef1α, rpb1, rpb2, csp1, and
β-tub1, are highly homologous to the corresponding segments
of the assembled genomes. MAT1-1-1, MAT1-2-1, β-tub1, csp1,
rpb1, and rpb2 are sensitive in distinguishing H. sinensis from
other fungal species. However, tef1α may be less sensitive and nrSSU and nrLSU are the least sensitive. For instance, EF468971
(nrSSU) and EF468827 (nrLSU) are highly homologous (97.4%-
99.5%) to the sequences of other taxa: Cordyceps multiaxialis (AJ309359), Cordyceps nepalensis (AJ309358), Hirsutella
leizhouensis (KY415580), Hirsutella rhossiliensis (MH872887,
NG_064109, EF546655), Ophiocordyceps acicularis (EF468805), Ophiocordyceps arborescens (NG_060238; AB968414), Ophiocordyceps
brunneanigra (MF614654, MF614653), Ophiocordyceps
geometridicola (MF614647, MF614648), Ophiocordyceps
macroacicularis (MH461122, NG_060239; AB968416;
MF614655), Ophiocordyceps multiperitheciata (NG_064462), Ophiocordyceps spataforae (MG831747).
The “ITS Pseudogene” Hypothesis for H. sinensis
Li et al. [27] proposed the “ITS pseudogene” hypothesis for the
AT-biased genotypes of O. sinensis, based on (1) the detection
of GC-biased Genotype #1 (H. sinensis) and AT-biased Genotype
#5 of O. sinensis in 8 of 15 cultures of a mono-ascospore
of natural C. sinensis and (2) the detection of the 5.8S cDNA
of Genotype #1, but not Genotype #5, in cDNA libraries constructed
from 2 H. sinensis cultures. However our study herein
found that the genome JAAVMX000000000 of strain IOZ07
contains 17 overlapped sequences of 5.8S genes, all of which
belong to GC-biased H. sinensis, Genotype #1 of O. sinensis.
The sequences of the AT-biased Genotypes #4-6, #15-17 of O.
sinensis resides not in the genome of H. sinensis but belong
to the genomes of independent fungi [8]. No 5.8S gene transcript
cDNA could be identified in the transcriptome assembly
GAGW00000000 of natural C. sinensis, indicating transcriptional
silencing of 5.8S genes consistently naturally occurred in all
colonized C. sinensis fungi. These findings provide solid evidence
contradicting against the “ITS pseudogene” hypothesis
for the AT-biased genotypes of O. sinensis fungi [6,10-11].
Intraspecific Variations of H. sinensis Genes
In contrast to the interspecies genetic variations among
the 17 genotypes of independent O. sinensis fungi [6-7,9-11,19,31,38,71], intraspecific variations at the genome and
transcriptome levels within the species H. sinensis have been
accessed herein through comprehensive cross-analysis among
the assembled and unassembled genome/mitogenome sequences,
the PCR-amplified sequences of the H. sinensis genes,
and the transcriptome sequences of H. sinensis strains and natural C. sinensis, while the taxonomic position of these H. sinensis strains (Genotype #1 of O. sinensis) was determined by the
authors of the original studies [4,15,24,26,28-29,33,35-36,39-41,49-52,53-60]. The cross-analysis did reveal intraspecific genetic
variations (similarities <97%) in a substantial number of
segment sequences of the H. sinensis genomes and transcriptomes,
although some technical errors might have occurred
during sequencing and assembling of the shotgun genome and
transcriptome sequences with using the second and third generations
of technologies.
Multigene analyses reported high genetic diversity of H. sinensis,
Genotype #1 of O. sinensis (a few of the diversified sequences
belonging to Genotype #3), isolated from natural C. sinensis specimens collected in southern Tibet or in western margin
areas and the central region of the Hengduan Mountains and lower genetic diversity of H. sinensis obtained from northern
and eastern margin areas of the Mountains [39-42,47]. Accordingly
these authors from the same research group proposed hypotheses
of “the center of origin” and “the fungal evolutionary
geography” for H. sinensis. In contrast to these hypotheses of
the geography-associated H. sinensis variations, Xiao et al. reported
large diversity in ISSR molecular marker polymorphism
among the 40 H. sinensis strains isolated from C. sinensis [44]
specimens collected from the same production area, Qingshashan
of Qinghai province, China. The cross-analysis of genome,
mitogenome, and transcriptome sequences presented herein
has verified intraspecific genetic variations in H. sinensis, Genotype
#1 of O. sinensis, which is distinct from the interspecies
variations among Genotypes #1-17 of O. sinensis [6-11-15,17-19,21-23,30].
Among the 33 marker genes (OSRC1-OSRC33) [41], we found
that the OSRC14, OSRC19, OSRC27, and OSRC32 contained
more mutant alleles in H. sinensis strains. In addition, 61 of the
254 unassembled genome sequences are genetically variable
when comparing to the assembled genome sequences of H.
sinensis strains. Many of OSRC marker genes and the unassembled
genome sequences were differentially transcribed in natural C. sinensis and strain L0106. Multiple overlapped sequences
of the 18S and 28S genes exist in the genomes of H. sinensis strains and multiple transcripts of the 18S and 28S genes were
found in natural C. sinensis. These overlapped gene sequences
contain scattered insertion/deletion, transition, and transversion
mutant alleles, some of which may cause translation interruptions
due to nonsense, frame shift, or missense mutations
of the genes. Some of the overlapped transcripts of the mutant
18S and 28S genes might be possibly derived from the fungi
colonized in natural C. sinensis [16,34].
A large number of H. sinensis strains have been arbitrarily
selected in the studies of O. sinensis fungi and natural C. sinensis insect-fungi complexes [4,8,12-13,15,17-19,21-22,26,28-30,33,35-36,39-40,44,72]. According to the ResearchGate
discussion with Dr. Nigel Hywel-Jones, Sung et al [4]. arbitrarily
selected strain EFCC7287 of H. sinensis (Genotype #1 of O.
sinensis) as the reference strain for fungus Cordyceps sinensis molecular taxonomy and nomenclature project and renamed
it to Ophiocordyceps sinensis. According to the sequences of
5 DNA loci, strain EFCC7287 is proven belonging to species H.
sinensis [6-7,71]. Sung et al. [4] therefore actually defined H.
sinensis (Genotype #1 of O. sinensis) as the sole anamorph of
teleomorph O. sinensis and did not expand the taxonomy-nomenclature
project to other 16 genotypes of O. sinensis that
belong to independent fungi, because of unavailability of pure
cultures of the mutant genotypes. Accordingly the rename of C. sinensis to O. sinensis is only restricted to H. sinensis, Genotype
#1 of O. sinensis. Moreover, many arbitrarily selected H.
sinensis strains have been used in industrial fermentation for
manufacture of commercial products, but studies are lacking
in exploring differences in pharmacological and toxicological
profiles between these strains and the type strain HMAS 55469
of H. sinensis [73]. Although Wei et al. [72] defined Cephalosporium
dongchongxiacaonis, Hirsutella hepiali, and Synnematium
sinensis as the synonyms of H. sinensis through molecular
systematic approaches, the H. sinensis type strain HMAS 55469
was not included in the study as the standard strain and the problematic molecular and bioinformatics methods used in the
study lead to uncertainty in the study conclusion [6,9,38,43-44]. Consequently the possible intra or interspecies variations
may need to be re-accessed at genome and transcriptome levels
among these so called “synonymic fungi” by the owners of
these fungal strains. Furthermore, direct comparisons in various
scientific disciplines (molecular mycology, genome/mitogenome,
transcriptome, proteome, metabolome and chemistry,
pharmacology, toxicology, etc.) are also absent between H.
sinensis strains that were submitted to government regulatory
bodies for product registration and the arbitrarily selected H.
sinensis strains for industrial use are often replaced later with
new strains because of unfortunate degeneration of the registered
strains with time. Our findings of significant intraspecific
genome and transcriptome variations in H. sinensis strains may
serve as an admonishment in academic and industrial use of H. sinensis strains and encourage scientists to establish a genome
standard for the H. sinensis type strain HMAS 55469 and
even transcriptome, proteome, and metabolome standards of
the type strain under various and standard fermentation conditions.
Conclusion
In conclusion, our findings present herein demonstrate alternative
transcriptions of many H. sinensis genes and apparent
intraspecific variations at the genome, mitogenome, and transcriptome
levels among H. sinensis strains. The results may
serve as a precaution for possible significant differences in
metabolome/chemical constituents, proteome, and pharmacology
between natural C. sinensis and mycelial fermentation
products of H. sinensis and possible alterations in the safety
profiles of H. sinensis-fermented products after arbitrarily exchanging H. sinensis strains for academic and industrial uses.
Inconsistent co-existence and alternative transcriptions of multiple H. sinensis mating-type genes in the H. sinensis strains and
natural C. sinensis are exactly the opposite of the homothallic
and pseudohomothallic mating previously hypothesized for H.
sinensis [24,26], and heterothallic mating may have to be considered.
Acknowledgements
This study was supported by grants from Natural Science Foundation
of Qinghai Province (2019-ZJ-967Q), the major science
and technology projects in Qinghai Province (2021-SF-A4), and
supported under the Cordyceps sinensis Industrial Development
Plan [QFGTZ (2020)491] by Qinghai Provincial Development
and Reform Commission.
Xiu-Zhang Li and Yu-Ling Li have contributed equally.
Conflict of Interests
None
REFERENCES
- Zhu JS, Halpern GM, Jones K (1998) The scientific rediscovery of an ancient Chinese herbal medicine: Cordyceps sinensis part I. J Altern Complement Med. 4(3):289−303.
[Crossref] [Google Scholar] [Pubmed]
- Zhu JS, Halpern GM, Jones K (1998) The scientific rediscovery of a precious ancient Chinese herbal regimen: Cordyceps sinensis part II. J Altern Complement Med. 4(4):429−457.
[Crossref] [Google Scholar] [Pubmed]
- Tan NZ, Barger JL, Zhang Y, Ferguson SB, Wu ZM, et al. (2011) The lifespan-extending effect of Cordyceps sinensis Cs-4 in normal mice and its molecular mechanisms. FASEB J. 25:599.1
[Crossref] [Google Scholar] [Research Gate]
- Sung GH, Hywel Jones NL, Sung JM, Luangsaard JJ, Shrestha B, et al. (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 57:5−59.
[Crossref] [Google Scholar] [Pubmed]
- Yao YS, Zhu JS (2016) Indiscriminate use of latin name for natural Cordyceps sinensis insect-fungi complex and multiple OphioCordyceps sinensis fungi. Zhongguo Zhong Yao Za Zhi 41:1361–1366.
[Crossref] [Google Scholar] [Pubmed]
- Zhu JS, Li YL (2017) A precious transitional Chinese medicine, Cordyceps sinensis: multiple heterogeneous OphioCordyceps sinensis in the insect-fungi complex. Lambert Academic Publishing, Germany.
[Crossref] [Google Scholar] [Research Gate]
- Zhu JS, Li YL, Yao YS, Xie WD (2018) Molecular evidence not supporting the use of Hirsutella sinensis strain EFCC7287 as the taxonomic standard for multiple OphioCordyceps sinensis fungi. Int J Mol Biol. 3:114−115.
[Crossref] [Google Scholar] [Research Gate]
- Xiao W, Yang JL, Zhu P, Cheng K, He H, et al. (2009) Non-support of species complex hypothesis of Cordyceps sinensis by targeted rDNA-ITS sequence analysis. Mycosystema 28:724−730.
[Crossref] [Google Scholar] [Research Gate]
- Li YL, Yao YS, Xie WD, Zhu JS (2016) The molecular heterogeneity of natural Cordyceps sinensis with multiple OphioCordyceps sinensis fungi challenges the anamorph-teleomorph connection hypotheses. Am J Biomed Sci. 8:123−159.
[Crossref] [Google Scholar] [Research Gate]
- Li XZ, Li YL, Yao YS, Xie WD, Zhu JS (2020) Further discussion with Li et al. (2013, 2019) regarding the “ITS pseudogene hypothesis” for OphioCordyceps sinensis. Mol Phylogenetics Evol. 146:106728.
[Crossref] [Google Scholar] [Pubmed]
- Zhu JS, Li YL, Yao YS, Xie WD (2019) The multiple genotypes of OphioCordyceps sinensis and the “ITS pseudogene” hypothesis. Mol Phylogenetics Evol. 139:106322.
[Crossref] [Google Scholar] [Pubmed]
- Engh IB (1999) Molecular phylogeny of the Cordyceps-Tolypocladium complex. Candidate scientific thesis, Department of Biology, University of Oslo, Oslo.
- Kinjo N, Zang M (2001) Morphological and phylogenetic studies on Cordyceps sinensis distributed in southwestern China. Mycoscience 42:567−574.
[Crossref] [Google Scholar] [Pubmed]
- Stensrud Ø, Schumacher T, Shalchian TK, Svegården IB, Kauserud H (2007) Accelerated nrDNA evolution and profound AT bias in the medicinal fungus Cordyceps sinensis. Mycol Res. 111:409−415.
[Crossref] [Google Scholar] [Pubmed]
- Zhang YJ, Xu LL, Zhang S, Liu XZ, An ZQ, et al. (2009) Genetic diversity of OphioCordyceps sinensis, a medicinal fungus endemic to the Tibetan Plateau: implications for its evolution and conservation. BMC Evolut Biol. 9:290.
[Crossref] [Google Scholar] [Pubmed]
- Zhang YJ, Sun BD, Zhang S, Wang M, Liu XZ, et al. (2010) Mycobiotal investigation of natural OphioCordyceps sinensis based on culture-dependent investigation. Mycosystema 29:518−527.
[Crossref] [Google Scholar] [Pubmed]
- Zhu JS, Gao L, Li XH, Yao YS, Zhao JQ (2010) Maturational alteration of oppositely orientated rDNA and differential proliferation of GC- and AT-biased genotypes of OphioCordyceps sinensis and Paecilomyces hepiali in natural Cordyceps sinensis. Am J Biomed Sci. 2:217−238.
[Crossref] [Google Scholar] [ResearchGate]
- Zhu JS, Zhao JG, Gao L, Li XH, Yao YS, et al. (2012) Dynamically altered expressions of at least 6 OphioCordyceps sinensis mutants in the stroma of Cordyceps sinensis. J. Fungal Res. 10:100−112.
[Google Scholar] [ResearchGate]
- Zhu JS, Yao YS, Li YL, Wu ZM, Liu X, et al. (2016) Hereditary variations with DNA segment substitution between fungi in the two types of Cordyceps sinensis ascospores suggest fungal chromosomal fusions. Proceedings of The 11th National Conference of Medicinal Mycology, Chengdu, China; pp.92−103.
[Crossref] [Google Scholar] [ResearchGate]
- Barseghyan GS, Holliday JC, Price TC, Madison LM, Wasser SP (2011) Growth and cultural-morphological characteristics of vegetative mycelia of medicinal caterpillar fungus OphioCordyceps sinensis G.H. Sung et al. (Ascomycetes) isolates from Tibetan Plateau (P.R. China). Int J Med Mushrooms. 13:565−581.
[Crossref] [Google Scholar] [Pubmed]
- Gao L, Li XH, Zhao JQ, Lu JH, Zhu JS (2011) Detection of multiple OphioCordyceps sinensis mutants in premature stroma of Cordyceps sinensis by MassARRAY SNP MALDI-TOF mass spectrum genotyping. Beijing Da Xue Xue Bao. 43(2):259−266.
[Crossref] [Google Scholar] [Pubmed]
- Gao L, Li XH, Zhao JQ, Lu JH, Zhao JG, et al. (2012) Maturation of Cordyceps sinensis associates with alterations of fungal expressions of multiple OphioCordyceps sinensis mutants with transition and transversion point mutations in stroma of C. sinensis. Beijing Da Xue Xue Bao. 44(3):454−463.
[Crossref] [Google Scholar] [Pubmed]
- Yao YS, Zhou YJ, Gao L, Lu JH, Wu ZM, et al. (2011) Dynamic alterations of the differential fungal expressions of OphioCordyceps sinensis and its mutant genotypes in stroma and caterpillar during maturation of natural Cordyceps sinensis. J Fungal Res. 9:37−49,53.
[Crossref] [Google Scholar] [Research Gate]
- Bushley KE, Li Y, Wang WJ, Wang XL, Jiao L, Spatafora JW, Yao YJ (2013) Isolation of the MAT1-1 mating type idiomorph and evidence for selfing in the Chinese medicinal fungus OphioCordyceps sinensis. Fungal Biol. 2013;117:599−610.
[Crossref] [Google Scholar] [Pubmed]
- Yao YS, Li YL, Wu ZM, Chen X, Liu X, et al. (2019) Hereditary variations of multiple OphioCordyceps sinensis fungi in two types of multicellular heterokaryotic Cordyceps sinensis ascospores. Proceedings of the 10th International Medicinal Mycological Congress, Nantong, Jiangsu, China, September 2019.
[Google Scholar] [Research Gate]
- Hu X, Zhang YJ, Xiao GH, Zheng P, Xia YL (2013) Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin Sci Bull. 58:2846−2854.
[Crossref] [Google Scholar] [Research Gate]
- Li Y, Jiao L, Yao YJ (2013) Non-concerted ITS evolution in fungi, as revealed from the important medicinal fungus OphioCordyceps sinensis. Mol Phylogenetics Evol. 68:373−379.
[Crossref] [Google Scholar] [Pubmed]
- Li Y, Hsiang T, Yang RH, Hu XD, Wang K, et al. (2016) Comparison of different sequencing and assembly strategies for a repeat-rich fungal genome, OphioCordyceps sinensis. J Microbiol Methods. 128:1−6.
[Crossref] [Google Scholar] [Pubmed]
- Li Y, Hu XD, Yang RH, Hsiang T, Wang K, et al. (2015) Complete mitochondrial genome of the medicinal fungus OphioCordyceps sinensis. Sci. Reports 5:13892.
[Crossref] [Google Scholar] [ResearchGate]
- Li YL, Yao YS, Zhang ZH, Xu HF, Liu X, et al. (2016c) Synergy of fungal complexes isolated from the intestines of Hepialus lagii larvae in increasing infection potency. J. Fungal Res. 14:96−112.
[CrossRef] [Google Scholar] [Research Gate]
- Li YL, Yao YS, Zhang ZH, Xie WD, Zhu JS (2018) Several genotypes of OphioCordyceps sinensis and Paecilomyces hepiali coexist in the two types of heterokaryotic Cordyceps sinensis ascospores. In Medicinal Fungi Section of China Edible Fungi Association. Proceedings of the 11th International Mycological Congress.
[Google Scholar] [Research Gate]
- Mao XM, Zhao SM, Cao L, Yan X, Han RX (2013) The morphology observation of OphioCordyceps sinensis from different origins. J Environ Entomol. 35:343−353.
[Crossref] [Google Scholar] [Research Gate]
- Liu ZQ, Lin S, Baker P, Wu LF, Wang XR, et al. (2015) Transcriptome sequencing and analysis of the entomopathogenic fungus Hirsutella sinensis isolated from OphioCordyceps sinensis. BMC Genome 16:106.
[CrossRef] [Google Scholar] [PubMed]
- Xia F, Liu Y, Shen GR, Guo LX, Zhou XW (2015) Investigation and analysis of microbiological communities in natural OphioCordyceps sinensis. Can. J. Microbiol. 61:104−111.
[CrossRef] [Google Scholar] [PubMed]
- Jin LQ, Xu ZW, Zhang B, Yi M, Weng CY (2020) Genome sequencing and analysis of fungus Hirsutella sinensis isolated from OphioCordyceps sinensis. AMB Expr. 10:105.
[CrossRef] [Google Scholar] [PubMed]
- Shu RH, Zhang JH, Meng Q, Zhang H, Zhou GL, et al. (2020) A new high-quality draft genome assembly of the Chinese Cordyceps OphioCordyceps sinensis. Genome Biol. Evol. 12(7):1074−1079.
[CrossRef] [Google Scholar] [PubMed]
- Quandt CA, Kepler RM, Gams W, Araújo JPM, Ban S, et al. (2014) Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5:121−134.
[CrossRef] [Google Scholar] [PubMed]
- Zhu JS, Wu JY (2015) Genetic heterogeneity of natural Cordyceps sinensis with co-existence of multiple fungi. Chin J Cell Biol. 37:284–298.
[Crossref] [Google Scholar] [ResearchGate]
- Zhang YJ, Bai FR, Zhang S, Liu XZ (2012) Determining novel molecular markers in the Chinese caterpillar fungus OphioCordyceps sinensis by screening a shotgun genomic library. Appl. Microbiol. Biotechnol. 95:1243−1251.
[CrossRef] [Google Scholar] [PubMed]
- Zhang YJ, Zhang S, Li YL, Ma SL, Wang CS, et al. (2014) Phylogeography and evolution of a fungal–insect association on the Tibetan Plateau. Mol. Ecol. 23:5337−5355.
[CrossRef] [Google Scholar] [PubMed]
- Zhang S (2013) Population Genetics of OphioCordyceps sinensis and its Evolutionary Relationship with Host Insects. Dissertation for Doctor’s Degree. Shanxi University, China.
- Li Y (2013) Epitypification, taxonomy and genetic diversity research on OphioCordyceps sinensis. Dissertation for Master’s Degree. Shanxi University, China.
- Ni LQ, Yao YS, Gao L, Wu ZM, Tan NZ, et al. (2014) Density-weighted algorithms for similarity computation and cluster tree construction in the RAPD analysis of natural Cordyceps sinensis. Am J BioMed Sci. 6(2):82−104.
[Crossref] [Google Scholar] [Researchgate]
- Xiao YY, Cao YP, Zhang L, Li CR (2014) Study on genetic diversity of anamorphic isolates from different sources of OphioCordyceps sinensis by ISSR. J Fungal Res. 12(3):154−159.
[Crossref] [Google Scholar] [ResearchGate]
- Yao YS, Gao L, Li YL, Ma SL, Wu ZM, et al. (2014) Amplicon density-weighted algorithms analyze dissimilarity and dynamic alterations of RAPD polymorphisms in the integrated micro-ecosystem Cordyceps sinensis. Beijing Da Xue Xue Bao. 46(4):618−628.
[Google Scholar] [PubMed]
- Debuchy R, Turgeon BG (2006) In Growth, Differentiation and Sexuality (eds. Ursula Kües & Reinhard Fischer). The Mycota. 293–323.
[Crossref] [Google Scholar] [Researchgate]
- Zhang S, Zhang YJ (2015) Molecular evolution of three protein-coding genes in the Chinese caterpillar fungus OphioCordyceps sinensis. Fungal China. 42(8):1549−1560.
[Google Scholar] [Researchgate]
- Kang XC, Hu LQ, Shen PY, Li R, Liu DB (2017) SMRT sequencing revealed mitogenome characteristics and mitogenome-wide DNA modification pattern in OphioCordyceps sinensis. Front Microbiol. 8:1422.
[Crossref] [Google Scholar] [PubMed]
- Chen JL, Chen YC, Yang SH, Ko YF, Chen SY (2009) Immunological alterations in lupus-prone autoimmune (NZB/NZW) F1 mice by mycelia Chinese medicinal fungus Cordyceps sinensis-induced redistributions of peripheral mononuclear T lymphocytes. Clin Exp Med. 9(4):277−284.
[Crossref] [Google Scholar] [PubMed]
- Quan QM, Wang QX, Zhou XL, Li S, Yang XL, et al. (2015) Comparative phylogenetic relationships of the caterpillar fungus OphioCordyceps sinensis and its host insects based on EF-1alpha sequnences. J Microbiol. 52(2):99-105.
[Crossref] [Google Scholar] [PubMed]
- Pandey A, Mohanty PS, Arya P (2010) Genetic diversity among the isolates of Cordyceps sinensis, a medicinal important fungi. GenBank. HM804956, HM804957, HM804958, HM804959, HM804960, HM804961, HM804962, HM804963, HM804964, HM804965, HM804966, HM804967, HM804968, HM804969, HM804970, HM804971, HM804972, HM804973, HM804974, HM804975, HM804976, HM804977, HM804978, HM804979, HM804980, HM804981, HM804982, HM804983, HM804984, HM804985, HM804986, HM804987, HM804988, HM804989, HM804990, HM804991, HM804992, HM804993, HM804994, HM804995
- Wen TC, Hyde K (2014) Fake OphioCordyceps sinensis finds its way to the Cordyceps market in China. GenBank. KJ364495, KJ364496, KJ364497.
[Google Scholar]
- Xiang L, Li Y, Zhu Y, Luo H, Li C, et al. (2014) Transcriptome analysis of the OphioCordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics 103:154−159.
[Crossref] [Google Scholar] [PubMed]
- Simmons DR, Kepler RM, Rehner SA, Groden E (2015) Phylogeny of Hirsutella species (Ophiocordycipitaceae) from the USA: remedying the paucity of Hirsutella sequence data. IMA Fungus 6(2):345−356.
[Crossref] [Google Scholar] [PubMed]
- Chan WH (2016) Authentication of DongChongXiaCao and abalone. GenBank.
[Crossref][Google Scholar] [PubMed]
- Chan WH, Ling KH, Chiu SW, Shaw PC, But PPH (2016) Molecular analyses of Cordyceps gunnii in China. J Food Drug Anal. 19
[Crossref] [ [Google Scholar] [Researchgate]
- Kang XC, Hu LQ, Shen PY, Li R, Liu DB (2017) SMRT sequencing revealed mitogenome characteristics and mitogenome-wide DNA modification pattern in OphioCordyceps sinensis. Front Microbiol. 8:1422.
[Crossref] [Google Scholar] [PubMed]
- Tian L, Hu B, Zhou H, Zhang W, Qu L, et al. (2016) RPB2 offer potential for increasing phylogenetic resolution in Cordyceps (Sordariomycetes: Clavicipitaceae) and its relevant taxa. Stud Mycol. 57:5-59.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Liu,YF, Yu H (2020) Tolypocladium reniformisporum sp. nov. and Tolypocladium cylindrosporum (Ophiocordycipitaceae, Hypocreales): two psychrotrophic fungi isolated from the same natural OphioCordyceps sinensis. Mycol Progress 21:199–214.
[Crossref] [Google Scholar] [Researchgate]
- Zhong X, Gu L, Wang HZ, Lian DH, Zheng YM, et al. (2018) Profile of OphioCordyceps sinensis transcriptome and differentially expressed genes in three different mycelia, sclerotium and fruiting body developmental stages. Fungal Biol. 122:943‒951.
[Crossref] [Google Scholar] [PubMed]
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, et al. (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569.
[Crossref] [Google Scholar] [PubMed]
- Xia E-H, Yang D-R, Jiang J-J, Zhang Q-J, Liu Y, et al. (2017) The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci Rep. 7:1806.
[Crossref] [Google Scholar] [PubMed]
- Liu Y-N, Jin Y-S, Liu Z, Han Y-N (2006)Cordyceps sinensis cDNA library construction and identification. Chin J Informat TCM 13(9):43−45.
[Google Scholar]
- Li X, Wang F, Liu Q, Li Q-P, Qian Z-M, et al. (2019) Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genomics 20:337.
[Crossref] [Google Scholar] [PubMed]
- Dong Y-Z, Zhang L-J, Wu Z-M, Gao L, Yao Y-S, et al. (2014) Altered proteomic polymorphism in the caterpillar body and stroma of natural Cordyceps sinensis during maturation. PLoS ONE. 9(10):e109083.
[Crossref] [Google Scholar] [PubMed]
- Zhang S, Zhang Y-J, Liu X-Z, Wen H-A, Wang M, et al. (2011) Cloning and analysis of the MAT1-2-1 gene from the traditional Chinese medicinal fungus Ophiocordyceps sinensis. Fungal China. 115:708−714.
[Crossref] [Google Scholar] [PubMed]
- Wilson AM, Wilken PM, van der Nest MA, Steenkamp ET, Wingfield MJ, et al. (2015) Homothallism: an umbrella term for describing diverse sexual behaviours. IMA Fungus. 6(1):207–214.
[Crossref] [Google Scholar] [PubMed]
- Zhang S-W, Cen K, Liu Y, Zhou X-W, Wang C-S (2018) Metatranscriptomics analysis of the fruiting caterpillar fungus collected from the Qinghai-Tibetan plateau. Sci Sinica Vitae. 48:562−570.
[Crossref] [Google Scholar]
- ZhaoJ-H Meng Q, Zhang H, Zhou G-L, et al. (2020) Transcriptomic analysis of the orchestrated molecular mechanisms underlying fruiting body initiation in Chinese cordyceps. Gene 763:145061.
[Crossref] [Google Scholar] [PubMed]
- Wei J-C, Wei X-L, Zheng W-F, Guo W, Liu R-D (2016) Species identification and component detection of Ophiocordyceps sinensis cultivated by modern industry. Mycosystema. 35:404–410.
[Crossref] [Google Scholar]
- Yao Y-S, Li Y-L, Zhang Z-H, Xie W-D, Zhu J-S (2018) Intraspecies genetic variations of Hirsutella sinensis (Genotype #1 of Ophiocordyceps sinensis) and transcriptome expression variations. Proceedings of the 11th International Mycological Congress, San Juan, Puerto Rico.
[Crossref] [Google Scholar] [Research Gate]
- Wei XL, Yin XC, Guo YL, Shen NY, Wei JC (2006) Analyses of molecular systematics on Cordyceps sinensis and its related taxa. Mycosystema. 25(2):192−202.
[Google Scholar]
- Liu X-J, Guo Y-L, Yu Y-X, Zeng W (1989) Isolation and identification of the anamorph of Cordyceps sinensis fungus. Acta Mycol Sinica. 8:35−40.
[Google Scholar] [Research Gate]
Citation: Li X-Z, Li Y-L, Yao Y-S, Zhu J-S (2022) Differential Expression of Hirsutella sinensis Genes and Intraspecific Genetic Variation
among H. sinensis Strains. Biochem Mol Bio J. 08:58.
Copyright: © Zhu J-S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License,
which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source
are credited.





