Review Article - (2016) Volume 2, Issue 1
Niculescu VF*
Kirschenweg 1, 86420 Diedorf, Germany
Corresponding Author:
Niculescu VF
Kirschenweg 1, 86420 Diedorf
Germany
E-mail: vladimir.niculescu@yahoo.com
Received date: Dec 21, 2015 Accepted date: Jan 20, 2016 Published date: Jan 22, 2016
Copyright: © 2016 Niculescu VF. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation: Niculescu VF. Developmental and Non Developmental Polyploidy in Xenic and Axenic Cultured Stem Cell Lines of Entamoeba invadens and E. histolytica. Insights Stem Cells. 2016, 2:1.
Secondary and tertiary progenitor cell lines of Entamoeba invadens (s-SRL and t-SRL lines) are unipotent. In controlled OCB cultures both cell lines proliferate asymmetrically giving rise to a minor cell fraction of MAS cells and a dominant cell fraction of MAT cells. While MAT cells re-enter mitotic cycle MAS cells switch into a developmental endopolyploid cell cycle (encystment/excystment cycle) that includes genome amplification, nuclear reprogramming and totipotency recovery. Each unipotent MAS cell gives rise to eight totipotent amoebulae that are the cells with the most differentiating potential. MAS cells form in cultures ATD cysts also observed in polyxenic isolates of E. histolytica and improperly called “spontaneous cysts”. In contrast, axenically grown E. histolytica proliferates by symmetric cell division and complete self-renewal; it gives rise to identical daughter cells. None of these cells enter the developmental endopolyploid cell cycle. Axenic stress hinders “spontaneous” encystment. However, we can distinguish in axenic cultures between minor and dominant subpopulations. While the dominant subpopulation cycled normally, cells belonging to the minor subpopulation undergo defective mitosis. They lose duplication fidelity and form multipolar spindles. As a result, the minor cell fraction becomes polyploid and multi-nucleated. Endoreplication by defective mitosis is reversible and has nothing to do with the process of encystment.
Keywords
Entamoeba invadens; Entamoeba histolytica; Encystment Competence; Endoreplication; Genome Reprogramming; Totipotency Recovery; Multipolar Mitosis; CLS
Abbreviations
Mat Cells: Mitotic Quiescent Cells Produced by the Tertiary T-Srl Cell Line; p-Srl: The Primary Stem Cell Line of Entamoeba; s-SRL: The Secondary Progenitor Stem Line; t-SRL: The Tertiary Progenitor Stem Line; OCB: Oxygen Consuming Bacteria; TD: Terminal Differentiation; ATD: Autonomous Terminal Differentiation; ITD: Induced Terminal Differentiation; CLS: Cyst Like Structures
Endopolyploidy: Goals and Purposes
Endopolyploidy is highly conserved in evolution and wide spread in eukaryotic cells. The origin and purpose of endoreplication, its function and relation to terminal differentiation and mitosis has been much discussed in the last years. In most cell types it evolves as a normal developmental process, linked to terminal differentiation or even part of it. Many authors considered polyploid cells arising from genome doublings as cells acquiring developmental features. Indeed endopolyploidy is in many organisms a late step of cell differentiation or a late aspect of the life cycle [1]. In conditions of stress and hypertension monoploid (haploid) or diploid cells develop higher levels of polyploidy (4N-64N) or a state of reversible hyperploidy [2-4]. Polyploidization was observed both in axenic stressed protist cultures [5,6-10] and in mammalian cell cultures [2,11,12] at early growth time points. The appearance of polyploid DNA contents in primary cell cultures suggests that endopolyploidization in culture is a common phenomenon analogous to natural ploidy states occurring in vivo [4].
A recent review focused on the differences observed during endoreplication and the multiple mechanisms used by monoploid or diploid cells to achieve a state of polyploidy [13]. A wide spread form of endoreplication is the developmentally controlled endocycle resulting in cells with a single polyploid nucleus that contains multiple copies of the normal DNA content. Endocycling is defined as a variant cell cycle in which DNA replicates but daughter cells do not form [1]. It consists of successive rounds of DNA synthesis (multiple S and G phases) occurring without chromosome condensation, mitosis, and cell division. Endocycling is a mitotic variant controlled by the same G1/S regulators that drive the normal mitotic cell cycle [13]. According to the most authors, polyploid cells are less suitable for classic mitosis [1]; Endocycling takes place only when classic mitosis is suppressed. However, after depolyploidization by reductive divisions progeny may return to mitotic cycles [3]. In contrast, endomitosis is an endoreplication process by incomplete mitosis. Endomitotic cells begin to condense chromosomes but do not segregate chromosomes to daughter cells. They enter a G1- like state and reenter S phase. Finally, re-replication is considered a more or less aberrant process of endoreplication, characterized by uncontrolled continuous re-initiation of DNA synthesis within a single S phase, leading to an indistinct DNA content without recognizable genome doublings.
It is evident that protist pathogens of the genus Entamoeba can proceed through different forms of endoreplication. One is the developmentally controlled endocycling linked with the process of terminal differentiation (encystment); it occurs both in E. invadens and E. histolytica. Developmental endopolyploization is part of an endopolyploid cell cycle that includes encystment and excystment processes [14]. The other is multipolar mitosis, the result of a defective mitosis by spindle defects occurring in axenic populations of E. histolytica experiencing environmental stress and nutrient deficiency [6-10]. Developmental endoreplication to mononuclear precursor cells containing eight whole genome copies is the precondition for terminal differentiation (encystment). For a better understanding of developmental and nondevelopmental polyploidy we first review encystment competence and cyst diversity in E. invadens and E. histolytica.
Encystment Competent Cells and Terminal Differentiation Pathways in Entamoeba Species
Recently we described in detail the primitive stem cell lineage (protolineage) hidden in the life cycle of E. invadens [15] which includes different processes of cell differentiation and cell line conversion, self renewal of stem cell lines by asymmetric cell division and complete self renewal by symmetric cell division. In our opinion, encystment is a process of terminal differentiation (TD), while ex-cystment is a process of de-differentiation generating undifferentiated reprogrammed totipotent amoebulae that give rise to primary stem cells. Encystment and cyst dissemination is a requirement for survival of pathogenic and free-living protists. Since each protist cell represents a single celled organism relying on itself, the wisdom above applies to every cell of the amoebic lineage regardless of its developmental stage, cell potency and cell cycle position. Most cells in cultures cannot encyst directly to ATD cysts. This is especially true for the dominant quiescent MAT cell fraction and its producer cells belonging to the t-SRL line that do not form ATD cysts. ATD cysts are produced only by the MAS cells exiting mitotic cell cycle. As a rule, only cells withdrawing from the mitotic cell cycle are capable of terminal differentiation [1,14].
Our understanding is that encystment is not only a mechanism of protist dissemination; at the same time it is a form of asexual reproduction, genomic reprogramming and totipotency recovery. Developmental processes lead in Entamoeba to eight totipotent amoebulae that re-start the stem cell lineage. Many innercyst cells are hatched out of cysts in the host or in OCB passages [15]. Thus, the key role of endopolyploidization and terminal differentiation in Entamoeba is totipotency recovery by genomic reprogramming. The reorganization from unipotent MAS precursor cells to totipotent amoebulae takes place via innercyst cell and metacyst cell development. The cyst wall protects the amoebic cell while reprogramming its genome.
Entamoeba invadens
As mentioned above, Entamoeba has a primitive stem cell lineage (protolineage). It starts directly from the totipotent amoebulae. The primary p-SRL stem cell line converts optionally to s-SRL or t-SRL lines and the s-SRL turn to t-SRL. All cell lines proliferate in OCB cultures by asymmetric cell division differentiating mitotic quiescent cells (primary MAP and tertiary MAT) and definitively arrested cells withdrawing mitotic cycle (secondary MAS cells) (Figure 1).
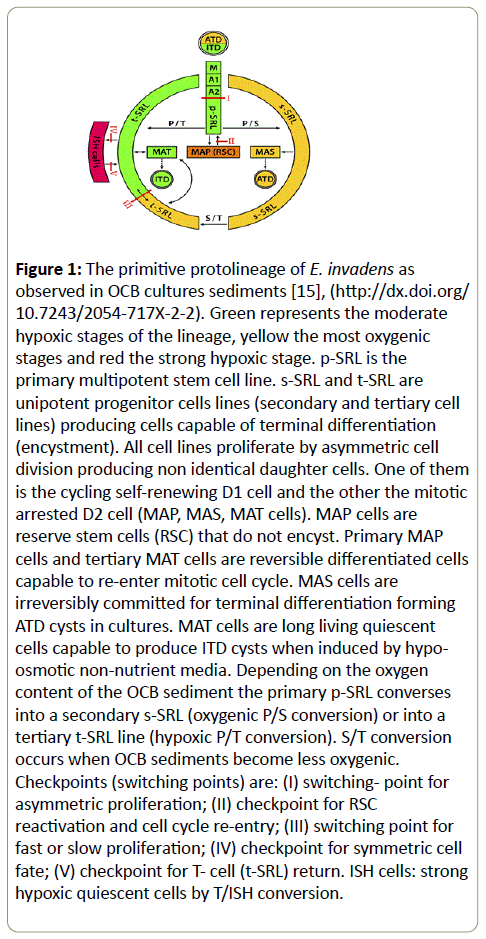
Figure 1: The primitive protolineage of E. invadens as observed in OCB cultures sediments [15], (https://dx.doi.org/ 10.7243/2054-717X-2-2). Green represents the moderate hypoxic stages of the lineage, yellow the most oxygenic stages and red the strong hypoxic stage. p-SRL is the primary multipotent stem cell line. s-SRL and t-SRL are unipotent progenitor cells lines (secondary and tertiary cell lines) producing cells capable of terminal differentiation (encystment). All cell lines proliferate by asymmetric cell division producing non identical daughter cells. One of them is the cycling self-renewing D1 cell and the other the mitotic arrested D2 cell (MAP, MAS, MAT cells). MAP cells are reserve stem cells (RSC) that do not encyst. Primary MAP cells and tertiary MAT cells are reversible differentiated cells capable to re-enter mitotic cell cycle. MAS cells are irreversibly committed for terminal differentiation forming ATD cysts in cultures. MAT cells are long living quiescent cells capable to produce ITD cysts when induced by hypoosmotic non-nutrient media. Depending on the oxygen content of the OCB sediment the primary p-SRL converses into a secondary s-SRL (oxygenic P/S conversion) or into a tertiary t-SRL line (hypoxic P/T conversion). S/T conversion occurs when OCB sediments become less oxygenic. Checkpoints (switching points) are: (I) switching- point for asymmetric proliferation; (II) checkpoint for RSC reactivation and cell cycle re-entry; (III) switching point for fast or slow proliferation; (IV) checkpoint for symmetric cell fate; (V) checkpoint for T- cell (t-SRL) return. ISH cells: strong hypoxic quiescent cells by T/ISH conversion.
Figure 1 represents Green represents the moderate hypoxic stages of the lineage, yellow the most oxygenic stages and red the strong hypoxic stage. p-SRL is the primary multipotent stem cell line. s-SRL and t-SRL are unipotent progenitor cells lines (secondary and tertiary cell lines) producing cells capable of terminal differentiation (encystment). All cell lines proliferate by asymmetric cell division producing non identical daughter cells. One of them is the cycling self-renewing D1 cell and the other the mitotic arrested D2 cell (MAP, MAS, MAT cells). MAP cells are reserve stem cells (RSC) that do not encyst. Primary MAP cells and tertiary MAT cells are reversible differentiated cells capable to re-enter mitotic cell cycle. MAS cells are irreversibly committed for terminal differentiation forming ATD cysts in cultures. MAT cells are long living quiescent cells capable to produce ITD cysts when induced by hypo-osmotic non-nutrient media. Depending on the oxygen content of the OCB sediment the primary p-SRL converses into a secondary s-SRL (oxygenic P/S conversion) or into a tertiary t- SRL line (hypoxic P/T conversion). S/T conversion occurs when OCB sediments become less oxygenic. Checkpoints (switching points) are: (I) switching- point for asymmetric proliferation; (II) checkpoint for RSC reactivation and cell cycle re-entry; (III) switching point for fast or slow proliferation; (IV) checkpoint for symmetric cell fate; (V) checkpoint for T- cell (t-SRL) return.
Multi-lined populations growing in OCB cultures show two distinct pathways of encystment. One is the autonomous encystment pathway (ATD) that gives rise to ATD cysts in cultures and the other is the induced encystment pathway (ITD) that gives rise to ITD cysts in encystment medium AaEM. ITD cysts occur in triple dose OCB sediments (3xOCB) immersed in the hypo-osmotic nutrient-free AaEM medium (Figure 1). Consequently, encystment competent cells are:
Secondary MAS (G0) cells encysting by ATD: MAS cells are mitotic arrested cells in the G0 state (definitively arrested cells) produced by the secondary s-SRL cell line. They are the single cell type of the Entamoeba lineage forming ATD cysts in complex cultures and nutrient-reach environments. These MAS precursor cells exit the mitotic cell cycle entering the developmental endopolyploid cycle. In our opinion this cell cycle variant includes endopolyploidization, encystment and excystment, de-polyploidisation and totipotency recovery. Repeated endocycling leads to eight whole genome copies. The polyploid precursor cell finishing endopolyploidisation is a mononucleated cell. Its conversion to cysts is dependant on the oxygen level in the culture. In optimal oxygenic conditions polyploid precursor cells form ATD cysts. In less optimal conditions they stop development and wait for oxygenation [16].
Early G1 cells (tertiary cells) encysting by ITD: The dominant tertiary subpopulation produced by the t-SRL stem cell line does not form ATD cysts in cultures, however, cells residing in the early G1 and the G0 state respond positively to encystment stimuli and form ITD cysts with a ratio of 1:1. Temporarily arrested MAT cells (G0 cells) re-enter the G1 phase at the same point as they exit it. In contrast to these early G1 cells all the other tertiary cells are mitotic cells involved in mitotic cycle progression and cannot leave it. As a rule cycling tertiary cells do not switch into the polyploid developmental cycle so long they are engaged in the mitotic cycle. The mitotic cycle suppresses terminal differentiation. However, encystment stimuli forces both G2 and G2/M cells to transition into the early G1 state thus becoming capable of terminal differentiation.
Thus, it becomes clear why cells of the late G1 and S phase exceeding the restriction point RP are not capable to endoreplicate and why G2/M cells generate cysts with a ratio of 1:2. Extrinsic encystment signals switch G2/M cells into a pre-encysting symmetric division. G2/M cells transfer encystment decision to both identical daughter cells and push them out of the mitotic cycle. Daughter cells encyst both from the early G1 state. These findings show the presence of a bifurcation switch- point that controls the fate of cells residing the early G1 phase: hypoosmotic non-nutrient conditions switch early G1 cells into the developmental endopolyploid cycle, while nutrient-rich media promotes G1 cell progression through the mitotic cell cycle. As reported in previous papers ATD encystment occurs in the first period of growth (t0-t30/ hrs) or in the pre-stationary/stationary growth phase (t96- t120/hrs) [15,17]. In the book “Anaerobiosis and Stemness - an evolutionary paradigm” published by Academic Press 2016, the authors Zoran Ivanovic and Marija Vlasky emphasize the key role of the O2 concentrations in eukaryotic differentiation/ terminal differentiation including protists as Entamoeba.
Entamoeba histolytica
It is generally accepted that axenic cultures of E. histolytica are not good for terminal differentiation (encystment). In contrast to polyxenic isolates and OCB cultures, axenic cultures do not allow amoebic cells to form ATD cysts. It is not clear why a significant number of cells at the end of the growth phase produce cyst-like structures (CLS) via intermediary cell forms (CLS precursor cells). CLS measure 8-20 μm in diameter [18]. They are sometimes bi- or trinucleated, however, they do not possess the characteristics of full normal cysts (thick protection wall, RNP bodies, four nuclei).
In polyxenic isolates E. histolytica form ATD cysts: As observed in cultures from clinical isolates - grown in Robinson’s old agar slant medium including rice starch and associate intestinal flora - E. histolytica possesses the capacity to form “spontaneous” cysts (ATD cysts) [19-23]. Moreover, long term populations of clinical isolates grown with commensal proliferating bacteria maintain for more than three years the whole developmental program forming cysts in cultures (G. Ehrenkaufer, U. Singh and C. G. Clark, unpublished data) [24,25].
Ehrenkaufer et al., rightly supposed that cell populations generated by complete (xenic) clinical isolates may contain all decisive developmental stages of E. histolytica [23]. The rare cysts observed by these authors are mononucleated juvenile cysts (ATD cysts) which have progressed sufficiently along the developmental pathway to form a true thick cyst wall and incipient chromatoid bodies. Moreover, cell populations generated by clinical isolates have high expression of developmentally regulated cyst-specific genes, homologous to encysting populations of E. invadens.
In other words, E. histolytica expresses encystment capacity if it is grown with living oxygen-consuming bacteria. There is no doubt that ATD cysts observed in polyxenic cultures are formed by MAS cells produced by the secondary stem cell line s-SRL. We assume that polyxenic cultures contain both stem cell lines (s-SRL and t-SRL) which proliferate by asymmetric cell division.
In axenic cultures E. histolytica form CLS: Said-Fernandez et al., reported that the HK9 strain of E. histolytica grown in 16 x 125 mm culture tubes filled with PEHPS or TPS-1 media generate a small proportion of viable, metabolically active specimens with an immature thin cyst wall [26]. These specimens are considered by the authors as “axenic cysts with an incipient (defective) cyst wall”. The reason are: (i) the mean cellular diameter of these specimens is of about 18.3 μm (1.78 times smaller than that of throphozoites); (ii) their cellular density is similar to that of true E. invadens cysts and less among that of axenic trophozoites and encysting precursor cells; (iii) the number of binucleated and tetranucleated CLS were two times higher than that of binucleated and tetranucleated trophozoites; (iv) the thin defective CLS wall contains chitin and binds calcofluor white M2R; (v) it is resistant to environmental insults as NaOH and hypothermia, however they are permeable for small molecules and completely deficient towards double-distilled water.
According to the authors in the first 3 days after inoculating axenic cultures the population increases by logarithmic growth (LG phase). It is a total of eight mitotic cell cycles and an average cell cycle of about 9 hrs. Within the next three days (days 4-6) cells enter the pre-stationary phase (DG phase) and growth gradually decreased. On day 7 the population reaches the stationary phase: 32.5% of the cells belong to a middle sized cell fraction of CLS precursors (“axenic precysts”) without a protective wall. Only 1.2% of this 32.5% fraction was tetranucleated however, the percentage of tetranucleated CLS was almost two-fold higher than that of tetranucleated trophozoites growing logarithmically. The authors considered these specimens as “viable axenic cysts”. During days 7 to 12 a massive vegetative cell death occurs (D phase). CLS reincubation with fresh PEHPS culture medium does not generate hatching or new vegetative populations.
Cyst wall structure: As described by Samuelson and Robbins [27] the cyst wall of Entamoeba is formed in two phases by a wattle and daub process. In the first wattle stage specific Jacob lectines are bound by the lectines of the plasma membrane and chitin fibrils are bound and cross-linked by the polyvalent Jacob lectines. During the second daub stage Jessie lectines - which are synthesized later - cells form the daub that makes the cyst wall of Entamoeba hard and impermeable for small molecules. We assume that CLS produce only the permeable wattle layer occurring as a rapid aid and counter measure against toxicity or nutrients consumption.
It is clear that specimens called by the authors “axenic cysts” are rather immature phenotypes representing an alternative product or a block in the differentiation process. Unfavourable environmental conditions switch part of the cell population to an incomplete differentiated phenotype; those cell that do not switch die [28-31]. Similar processes are known in other eukaryotic cell systems where differentiation programs and massive cell death (apoptosis) are tightly linked together [32]. In these systems cell death occurs preferentially in the G2/M phase while terminal differentiation occurs when cells exit the mitotic cell cycle or arrest in G1 [32]. This could be an explanation for why a minor part of the population produced specimens protected by an incomplete cell wall layer (consisting of Jacob lectines and chitin fibrils), while the dominant part of the population did not.
Methods improving CLS formation:
Preconditioning: Amoebic populations of E. histolytica grown in a more hypoxic TYI-S-33 culture medium preconditioned by bacterial metabolites give rise to a high yield of surviving CLS similar to the specimens observed in PEHPS cultures [18,26,33].
Oxygenation by H2O2: Aguilar -Diaz et al., cultured amoebae of the HMI-IMSS strain in culture flask containing 50 ml medium for 72 hrs [33]. They added H2O2 to a final concentration between 2-8 mM, re-incubating the culture samples for 2-24 hrs at 37°C. The greatest CLS conversion rates (~30%) occur in cultures supplemented with 4 mM H2O2 and then re-incubated at 37°C for 6 hrs. The viability of the amoebic population treated with 4 mM H2O2 decreased in time. It decreases to 90% (after 2 hrs), 65% after 6 hrs and 10% after 24 hrs. About 35% of the induced CLS possessed two nuclei and 15% possessed three nuclei.
CO2 treatment: Barron-Gonzales et al., used the two phase hypoxic culture method of Morales-Vallarta et al., to culture amoebae of the HMI-IMSS strain in TYI-S-33 medium enriched by bacterial metabolites [18,34]. The first growth phase occurs in 18 × 150 mm tubes with 10 ml medium (72 hrs), the second phase in Erlenmayer flasks containing 30 ml medium (96 hrs). After inoculation, cultures were treated by CO2 gas for 2 min (first culture) and 6 min (second culture) respectively. 72 hrs after innoculation, the first amoebic populations were harvested and centrifuged, then washed and transferred in Erlenmeyer flask containing fresh medium without glucose. Flask medium was supplemented with sterile bacterial metabolites from E. coli and Enterococcus faecalis. PBS washing and amoebic transfer into culture flasks meant a period of aeration between the two hypoxic growth phases.
96 hrs after starting the second hypoxic culture, 92% of the cells are CLS specimens. However, the data do not reveal the number of doublings and the duration of amoebic cell cycle. It is not clear, if a massive cell death takes place in the stationary growth phase (72-96 hrs) or not. This would be essential to know the origin and the actual rate of CLS formation as referred to the total number of trophozoites at the end of logarithmic growth phase. CLS with more than one nucleus are in our opinion not necessarily mature cyst specimens as suggested by the authors [18]. They might rather be a semidifferentiated phenotype starting from multi-nucleated polyploids after multipolar mitosis.
Hypoosmotic pressure: an inducer for hidden ITD encystment?
Said-Fernandez et al., investigate the effect of a hypoosmotic extremely diluted culture media (55.6 mOsm/kg) on tubes containing CLS (“axenic cysts”) and CLS-precursors (“encysting amoebae”) which were previously incubated for 3-6 days in PEHPS and TPS-1 culture medium. Specimens from the logarithmic (day 3) and pre-stationary growth phase (day 6) were sedimented by gentle centrifugation, then exposed to the hypoosmotic shock (24 hrs/4°C) and then re-incubated with fresh culture medium. Tubes were then re-cultured for 7 days in fresh medium at 36°C and observed daily by inverted microscopy [26].
7 days after re-incubation, some of the hypoosmotically treated samples containing the CLS/CLS-precursor fraction gave rise to a new vegetative population, while control samples (not hypoosmotically treated) did not. Samples of the 3 days old PEHPS cultures free of CLS/CLS-precursor cells and 6 day old TPS-1 cultures containing CLS/CLS-precursor cells are not capable of generating vegetative populations when recultured. Only the 6 day old PEHPS culture containing CLS/CLSprecursors can do this. Hypoosmotic shocks lasting more than 24 hrs prevented vegetative cells restoration.
We believe that hypoosmotic treatments shorter as 24 hrs and a more rapid exposure to fresh culture media may be beneficial for vegetative regeneration. Similar to the encystment of E. invadens that begins in AaEM encystment medium (1-3 hrs) and continues in nutrient-rich media for 24 hrs the CLS/CLS-precursor fraction would lead to ITD cysts [16]. These cysts probably remained unobserved by Said-Fernandez et al. [26]. In other words we believe that the new vegetative population starts by hypoosmotic induced ITD cysts that excysts in re-culture.
In our opinion, hypoosmotic treatment and medium substitution by PEHPS implies oxygenation of the CLS/CLSprecursor fraction. Submersion in hypoosmotic nutrient-poor media favors encystment [15,35]. We suppose, oxygenation and hypoosmotic shock are effective inducers for ITD encystment in E. histolytica.
There is currently no evidence which cell type was capable of forming true ITD. In any case: hypoosmotic treatment of pre-stationary PEHPS populations containing CLS/CLSprecursor fractions could be a consistent method for producing true cysts of E. histolytica. This technique is quite similar to the method introduced by us 1974 to encyst whole populations of E. invadens by oxygenation and hypoosmotic treatment in nutrient-free encystment medium [35]. However, the method of Said-Fernandez et al., applies only to cells from the stationary growth phase (6 day old culture) containing CLS/ CLS-precursor cells and has no impact on younger logarithmic vegetative cultures (3 day old cultures) lacking the CLS/CLSprecursor fraction [26].
The Irreversibility of Developmental Endopolyploidization
Each cell switching in the developmental endopolyploid cycle (MAS cells in nutrient-rich media and early G1 cells in hypoosmotic nutrient-free media) passes a point of no return. Consequently, developmental endopolyploidization is irreversible and developmental endopolyploid cells do not revert into mitotic proliferative cells.
MAS cells are precursor cells of ATD encystment. They switch in OCB and xenic cultures into the endopolyploid cell cycle. MAS precursors amplified their genome to the eight genome copies needed for encystment. The resulting polyploid precursor cells are mononucleated. After a phase of cell maturation the mononucleated mature polyploid precursor cells cross the checkpoint of terminal differentiation (TD checkpoint) forming juvenile ATD cysts. MAS cell development is an oxygenic process that needs appropriate environments [15,17]. Terminal differentiation continues by cyst maturation and partial genome segregation to four diploid nuclei. The final product of maturation is the tetranucleated mature cyst. The endoployploid cycle finishes by excystment and final genome segregation into eight monoploid (mononucleated) amoebulae (Figure 2).
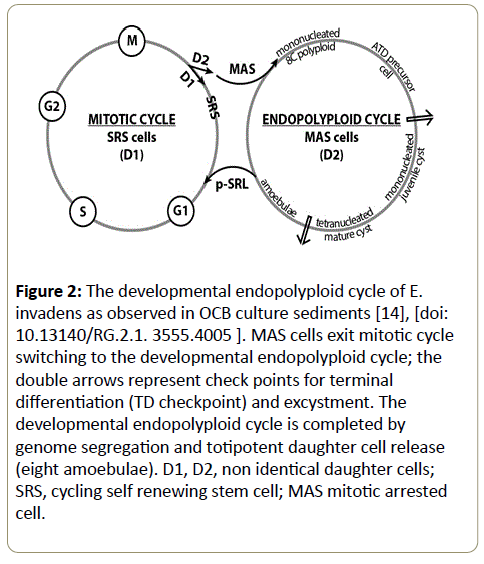
Figure 2: The developmental endopolyploid cycle of E. invadens as observed in OCB culture sediments [14], [doi: 10.13140/RG.2.1. 3555.4005 ]. MAS cells exit mitotic cycle switching to the developmental endopolyploid cycle; the double arrows represent check points for terminal differentiation (TD checkpoint) and excystment. The developmental endopolyploid cycle is completed by genome segregation and totipotent daughter cell release (eight amoebulae). D1, D2, non identical daughter cells; SRS, cycling self renewing stem cell; MAS mitotic arrested cell.
Quiescent MAT cells in cultures do not exit the mitotic cycle. After passaging to subcultures they continue proliferation by asymmetric cell division forming cycling cells that replenish the t-SRL stem cell lines and new quiescent MAT cells. However, appropriate stimuli from hypoosmotic nutrient-free media revert MAT/G0 cells into G1 state and promote G1 cells into the developmental endopolyploid cycle [15,17]. G2/M cells were induced in hypoosmotic nutrient- free media to symmetric cell division. These cells give rise to identical daughter cells in the early G1 state. Both identical daughter cells enter the developmental cycle and form ITD cysts.
Cells entering the developmental endopolyploid cycle do not revert to mitotic cycle; their ploidy is irreversible. From a cell biological point of view, the developmental cycle is a mechanism for totipotency reprogramming and lineage rejuvenation. Unipotent cells, such as MAS and MAT cells and undifferentiated cells in G1 state, generate totipotent amoebulae capable to start a new amoebic stem cell lineage.
Non-developmental Polyploidy is Reversible
Cultures of Entamoebae are multilined and geneticly heterogenous [15]. Several reports in the literature show that axenic populations of E. histolytica contain a minor cell fraction of about 11-15% cells cycling by defective multipolar mitosis and a dominant subpopulation ~90%, free of mitotic disorders [10,36]. Both cell fractions proliferate in axenic cultures by symmetric division giving rise to identical cycling cells (complete self-renewal).
We suppose that the minor axenic cell fraction represents the s-SRL stem cell line switched in the complete self renewal mode. Consequently, none of the axenic grown cells switch to the developmental endoployploid cycle. Thus, axenic stress hinders asymmetric cell fate, mitotic arrest, cell differentiation, mitotic cycle exit and “spontaneous” (ATD) encystment. Endoreplication by defective multipolar mitosis observed in the minor cell fraction has nothing to do with the process of encystment. It is a non-developmental transient polyploidy induced by spindle defects.
Defective polyploids may be easily induced to ploidy reversal [10]. They undergo successful de-polyploidization leading to mitotically cycling survivors that restart endopolyploidization by multipolar mitosis. Analogous processes are known in mammalian stem cell systems and rejuvenated cancer cells [37,38].
Multipolar Mitosis in E. histolytica
Mitotic disorders begin by abnormal centrosome amplification that give rise to abnormal centrosome number (3-4 centrosomes instead of normal duplication fidelity) and dictates the formation of multiple spindle poles. Multipolar spindles lead to multinucleated cells. However, these multinucleated cells are cycling mitotic cells and not ATD precursor cells (definitively mitotic arrested cells). Consequently, the nuclear DNA content of axenic cultures is higher than those of xenic cultures [6-10,36,39].
Synchronization experiments using serum deprivation and serum re-addition performed by Mukherjee et al., revealed that the nuclear cycle of this minor cell fraction of 10-20% lasted of about 9 hrs (t1-t9) [10]. The authors found the lowest DNA content at t1 and t10; 10-20% of nuclei were polyploid accumulating DNA contents greater than 4N; the number of binucleated cells increased between t0 and t9 progressively, however not exceeding 19% cells (average value ~16%); About 12% of the cells formed multitubular spindles (MT assemblies) between t4 and t8. Their number is halved at t12. The increase in binucleated cells corresponds to the decrease of MT structures and the reduction of the nuclear DNA content.
Improper environmental conditions cause often mitotic disorders in eukaryotic cells. Mitotic and cytokinetic disfunctions generate high rates of segregation errors by supernumerary centrosomes and MTOCs (microtubule organizing centers) [40]. In consequence, polyploid cells undergo multipolar division or cell fusion [41].
Depolyploidization by Promeiotic Cell Division
The asynchrony observed in axenic culture of E. histolytica between genome segregation and cell division can have different origins. Environmental conditions compromising the fidelity of the inherited genome activated checkpoint control pathways that delay cells to allow them to finish a mitotic cell cycle [41]. In various systems cell exit from mitosis is controlled by spindle assembly check points and specific proteins that allow chromatid separation. However, a broad class of agents induces the loss of cell regulation cycle [42-47]. Other cells conserved mechanisms of ordered polyploid genome disassembly by promeiotic pathways that initiate depolyploidization [41]. One other pathway of disregulation is the abnormal centrosome amplification which creates multipolar spindles [42]. A normal cell has two centromeres at mitosis, which dictate the formation of two spindle poles. More duplication events lead to multipolar spindles and disordered genome segregation. Elimination of regulatory mechanisms that governs centrosome duplication leads to centrosome amplification and multipolar mitosis [48,49].
According to the observations of E. histolytica researchers axenic mitotic defective cells tend to hyperploidization (16N genome) and multinuclearity by asymmetric nuclear division [6-10]. Tripolar and tetrapolar spindles were observed [10]. Some of the nuclear segregation patterns are not surprising; they remind one of the segregation patterns observed in amoebulae genesis by Dobell and Dutta [19,50].
Reductive nuclear division is common in cells possessing intrinsic control mechanisms for genome segregation [51]. Such mechanisms are activated in the developmental cycle of Entamoeba (s-SRL lineage) during innercyst cell maturation and genome segregation. The eight monoploid amoebulae originate by intra- and metacystic reductive nuclear division.
The ability of polyploid cells to undergo depolyploidization is wide spread in mammalian embryo cells and cancer cells, present as an internal control mechanism for genome segregation [41]. Molecular analysis showed meiosis-specific genes that were activated during the depolyploidization process. These gene products allow reorganization of the genetic material giving rise to viable descendants. Depolyploidizating cells acquire pseudo-meiotic properties switching from a promitotic to a promeiotic division pattern. In this way descendants stabilize genetic alterations and acquire a stable growth state.
Meiosis-specific genes DMC1, SPO11 and MND1 are reported for E. histolytica [52]. Singh et al., found increased rates of genetic recombination in Entamoeba under stress conditions and during encystment, suggesting that Entamoeba might engage in genetic recombination at different stages of their life cycle. Molecular mechanisms of depolyploidization assure polyploidy reversal in both endopolyploid and defective mitotic cycle of Entamoeba [53].
Endopolyploidy, Genome Reprogramming and Totipotency Recovery
Restricted developmental reprogramming vs. reversed pluripotential reprogramming
According to Gurdon and Melton reprogramming is “a process by which the differentiation state of a cell is changed to that of another state” [54]. During mammalian embryogenesis developmentally plastic stem cells become restricted to particular highly stable cellular lineages; differentiated somatic cells very rarely changed from one differentiated state to another [55]. In the last years the differentiated state of a somatic cell has been reversed experimentally to pluripotency by nuclear transfer, cell fusion, pluripotent cells extracts, transcription factor overexpression or transdifferentiation [55-58].
Genome reprogramming is a natural unidirectional process characterized by distinct epigenetic modifications [59,60]. It involves resetting epigenetic mechanisms (epigenetic marks) that maintain stable gene expression, such as DNA methylation, histone methylation, phosphorylation and deacethylation that represent key events in differentiation processes [55,61-63]. Epigenetic changes alter gene expression and chromatin organization without changing the DNA sequence. Genome reprogramming by “epigenetic patterns” occurs during development and differentiation as a response to intrinsic and environmental stimuli (https://www. rocklandinc. com/Epigenetic-marks-cell-cycle-control.aspx).
Successful nuclear reprogramming involves fundamental changes in cellular functions such as DNA replication and nuclear organisation [55]. The authors consider “the identity of any cell to be determined by the expression of the lineage specific genes that confer cellular identity (cellular phenotype). In order to change a particular identity a cell has to turn on new lineage specific genes and turn off the old ones”. The most common structural rearrangements play a role in the cell cycle control, creating a G1 checkpoint [64].
According to Jaenisch and Young the gene expression of pluripotent cells is the product of regulation by specific factors such as chromatin- modifying enzymes and signal transduction factors. Multiple interacting proteins likely coordinate and control pluripotency as developmental regulators [65,66].
Multipotency restriction and totipotency recovery in Entamoeba
In single celled eukaryotes as E. invadens the epigenetic restricted unipotent cell state of secondary and tertiary cells is not irreversibly fixed and can be reprogrammed to an undifferentiated totipotent cell state that starts a new cycle. Genome reprogramming occurs in all cells that enter and progress through the developmental endopolyploid cell cycle; it concerns both secondary MAS cells and early tertiary G1 cells. The developmental endopolyploid cell cycle of Entamoeba likely contains a sequence of epigenetic events such as chromatin modifications or changes in DNA methylation controlled by intracellular reprogramming factors.
The mechanisms of protist reprogramming by genome amplification and subsequent segregation are largely unknown. One could assume that the most common epigenetic rearrangements occur in the late G1 phase before genome amplification. However, chromatin changes associated with reprogramming appear to happen progressively from one state to the other [65]. It would be conceivable that epigenetic rearrangements can also occur in the phase of polyploid precursor maturation [16].
After ex-cystment, totipotent amoebulae give rise to the multipotent p-SRL stem cell line. Similarly to pluripotent cells, the multipotent primary cells of E. invadens are poised to rapidly differentiation in culture. Molecular circuitry allows their differentiation into the more specialized progenitor cell lines s-SRL and t-SRL in response to particular signalling cues [15]; unipotent MAS cell produced by the s-SRL line, in culture, form ATD cysts. Similarly, spermatogonial stem cells (that are known to be unipotent in vivo) give rise in cultures to distinct ES-like cells (ES, embryonal stem cells) [67]. The authors talk about culture-induced reprogramming. Pluripotency recovery evidently activates ancestral mechanisms of the protist life cycle.
Endopolyploidy and stemness
Genome reprogramming and endoreplication seemed to be linked together. In some cases somatic stem cells could be reprogrammed to pluripotency by modifying cell culture conditions and the resulting iPS cells are polyploidy [60]. Other cells can revert through induced polyploidy and reprogramming for totipotency even from terminal senescence [68].
Authors studying the immortality of cancer cells after genotoxic treatment, found evidence suggesting polyploidy as a link between genotoxic stress, cellular senescense, cancer development and cancer renewal after treatment [69-71]. Erenpreisa and Cragg hypothesized the existence of a “cancer cell life cycle” presuming the activation of germline-like mechanisms of unicellular organisms (via polyploidy induction and ploidy reverse) and the subsequent appearance of a stemcell- like phenotype; this reversible polyploidy activates meiotic genes also [37].
Conclusions
Developmental and non-developmental ploidy of Entamoeba is fundamentally different. In contrast to nondevelopmental ploidy, developmental ploidy is irreversible; it precedes terminal differentiation and is part of an endopolyploidization-depolyploidization cell cycle outside the mitotic cell cycle. This developmental endopolyploid cycle is a developmental cycle hidden in the processes of encystment and excystment. It includes genomic reprogramming and totipotency recovery giving rise to totipotent daughter cells (amoebulae) capable of starting a renewed life cycle comprising a new stem cell lineage. During the developmental endopolyploid cell cycle chromatin changes associated with reprogramming appear to happen progressively from one cell state to the other. It would be conceivable that epigenetic rearrangements occur both in the phases of endoreplication and polyploid precursor maturation.
Two cell types are capable of exiting the mitotic cell cycle and forming cysts by terminal differentiation: the secondary MAS cells and the postmitotic (just born) G1 cells before entering the mitotic cycle. MAS cells enter the endopolyploid cell cycle forming ATD precursor cells by intrinsic mechanisms. In optimal growth and environmental conditions mature precursor cells pass through the checkpoint of terminal differentiation and form ATD cysts. Older G1 cells must be induced to exit the mitotic cell cycle by extrinsic stimuli. These cells enter the developmental cycle and form ITD cysts.
Genetic heterogeneity observed in cultures of E. histolytica is caused by the simultaneous appearance of several stem cell lines in cultures. Long term amoebic populations are usually multi-lined. They consist of a minor secondary s-SRL and a dominant tertiary t-SRL forming minor and dominant subpopulations. In contrast to the OCB and polyxenic cultures, axenic populations proliferate by symmetric cell division (complete self-renewal). While the dominant cell line (t-SRL) performed an apparently normal mitotic proliferation, the minor cell line (s-SRL) has difficulties to preserve mitotic fidelity. The result is a genetically heterogeneous population containing multinucleated cells with a ploidy of 1N to 14N. 48 hrs after starting axenic cultures consists up to 92% monoploids and 15-20% polyploids greater as 6N.
Extrinsic cues (axenic stress) disturb the mitotic fidelity of the minor cell line. Defective multipolar mitosis by spindle insults leads to non-developmental transient polyploidy and multinucleated polyploids unrelated to terminal differentiation and encystment. These cells are not the normal precursors of tetranucleate cysts (“precysts”) as suggested in the past. Similar disorders occur neither in the dominant cell line nor in the minor cell line grown under xenic conditions. All xenic cells perform normal mitosis.
In light of these findings, in Entamoeba, one cannot speak of inherent amoebic features decoupling nuclear division from cell divisions or missing cell cycle check points as has happened in the past.
Acknowledgement
The author expresses his gratitude to Dr. Dennis Thomas (native English speaker) for reading of the manuscript and excellent support.