Keywords
Volunteer computing, De novo assembly, Virtual super computer
Introduction
Volunteer computing is the process
of using computing resources of people,
who are willing to donate their computing resource (called as volunteers) to compute or
store huge data. When we browse internet,
most of the computer resources will be idle. On the other hand there are many
applications mostly scientific which are
lying around for the lack of computing
resources. Many scientific applications
started using these idle resources by
convincing the owners of the resource to
owe them, and to come up with a new way
of computing called as volunteer computing.
Search for extraterrestrial intelligence
(SETI) SETI@home [5], a scientific area
whose goal is to detect intelligent life
outside Earth, was the first application that
started to use idle computing resources from
all over the world to form the largest
computation power of near 70Teraflops.
Even today it is the largest project with more
than million computers supporting the
project.
Berkeley Open Infrastructure for
Network Computing (BOINC) [6] is an open
source middleware designed and developed
to support applications that require high
computational power, data storage or both
such as SETI@home, Climateprediction [7],
Folding@home [1]. It helps the scientists to
create applications that use public
computing resources. It also enables the
volunteers to support multiple projects at the
same time and a way to manage their
resources contribution. It is designed in such
a way that it is not susceptible to a single
node failure and can be used for a wide
range of applications.
Although volunteer computing as an
alternate form of grid computing, is enabling
the creation of largest computation units in
the world. Many applications are not yet
willing to adopt his method of computation
because of their security concerns to send
their valuable data on internet. This is also
the reason for many scientists for not using
many computing technologies that use
internet such as cloud.
Some of these large scale
applications do not require the computing
power to be gathered from whole world. Many organizations such as colleges,
training institutes possess large collections
of desktop systems that stay powered on but
idle most of the time in a day. These
laboratories can be used to create a cluster of
systems that form a grid of volunteer
computers. It eliminates the worry about the
security of the data as it stays completely in
our control and it also doesn’t need any
expensive internet connections.
In this paper, the creation of a virtual
super computer in the laboratories of the
college to solve the computational and data
intensive application of “Assembling the
Reads of Genome” using proven de novo
assembly techniques for large datasets has
been discussed.
This paper is organized as follows,
section II gives the overview of the
application, section III contains the
architecture using BOINC, section IV
explains an existing algorithm used in the
application and the reasons for selecting the
same, section V presents the methodology of
how legacy applications and algorithms can
be easily converted into BOINC compatible
applications, section 6 concludes the paper
and gives future enhancements.
Overview - Dna Sequencing
Deoxyribonucleic acid (DNA)
sequencing is the process of using any
method to determine the correct order
of nucleotides bases Adenine (A), Guanine
(G), Cytosine (C) and Thymine (T) within a
strand of DNA molecule [16]. These bases form
pairs to form the double helix structure of the
DNA and usually called as base pairs. It can
be used to sequence a small genome or the
whole genome of any organism. It is used in a
wide variety of fields such as anthropology,
genetics, biotechnology, and forensic
sciences. Due to huge size of the DNA
(usually contains of millions of base pairs) it
is not possible to identify the sequence of all
nucleotides at a single shot.
This process can be best compared
with relevant to exact ordering of words in a
page that is shredded into small pieces. Some
of the problems [2] that make this process more
difficult are that some of the words may be
missing or some words may be duplicated.
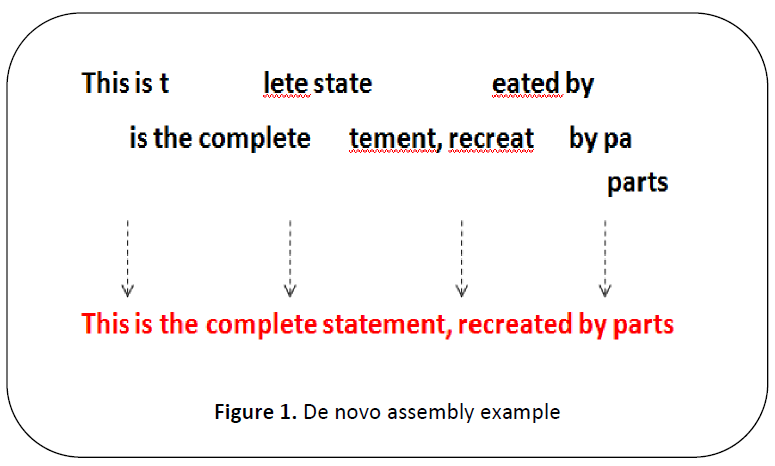
The usual solution is to assemble the words to
make a sentence as shown in Figure 1 and
assembling those sentences to get the whole
page.
Figure 1: De novo assembly example
Similarly in DNA sequencing the
words are called as reads and they are
assembled back to sentences called as contigs
and assembled to pages similar to
supercontigs (also called as scaffoldings) as
explained in Figure 1.This process is called as
de novo assembly [23]. This de novo assembly is
primarily used in assembling the reads
produced by next generation sequencers.
These assembly algorithms use graph
based techniques to find the reads with high
probability of becoming neighbors. Graph
based assembly techniques are usually
classified into two sub classes, the Overlap-
Layout-Consensus (OLC) [18] method and the
De Bruijn Graph (DBG) [14] method. Both these
methods assemble the whole genome
sequence by exploiting overlap information
about reads that conforms to the Lander-
Waterman Model [10]. In terms of space and
time complexity, OLC algorithms are
preferable for low-coverage long reads and
DBG algorithms are the better choice for
high-coverage short read and large genome
assembly, further comparisons of the both can
be found at [25].
OLC method is based on an overlap
graph. Generally speaking, this method works
in three steps. First, overlaps (O) among all
the Reads are found. Following that, OLC
method creates a layout (L) of all the reads
and overlaps the information on a graph and
finally the consensus (C) sequence is inferred
from the Multiple Sequence Alignments
(MSA).
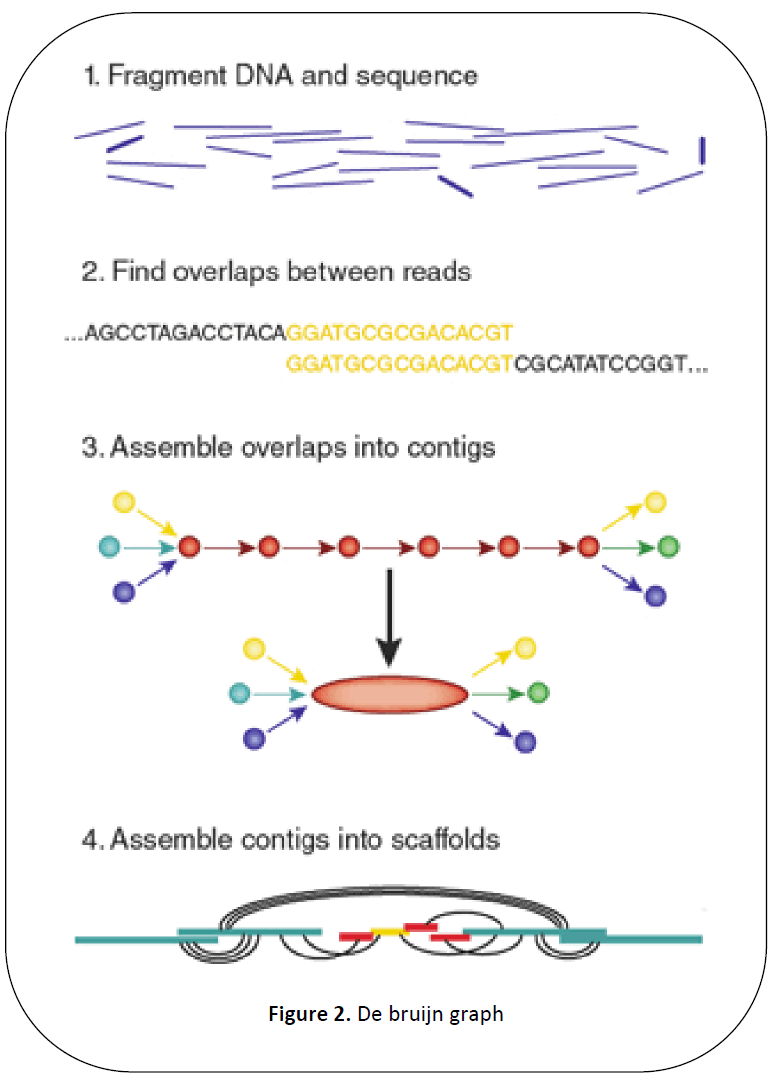
The DBG works by first chopping
reads into a set of much shorter k-mers and
then making use of these k-mers and finally
inferring the whole genome sequence from
the DBG.
As explained in Figure 2 the DBG is
created by a set of contigs and scaffolding.
Then the DBG is read through beginning with
a short read to get the entire sequence back.
Figure 2: De bruijn graph
BOINC grid middle-ware architecture
Although there are many grid
middleware architectures such as condor [17],
GLOBUS [15], CFD [22], BOINC has been
selected as the optimal solution for many
scientific applications because it is scalable
and has the ability of high-performance task
distribution, high fault tolerance
performance [21]. The other methods require
more administration compared to BOINC for
volunteer computing and have less check
pointing mechanism.
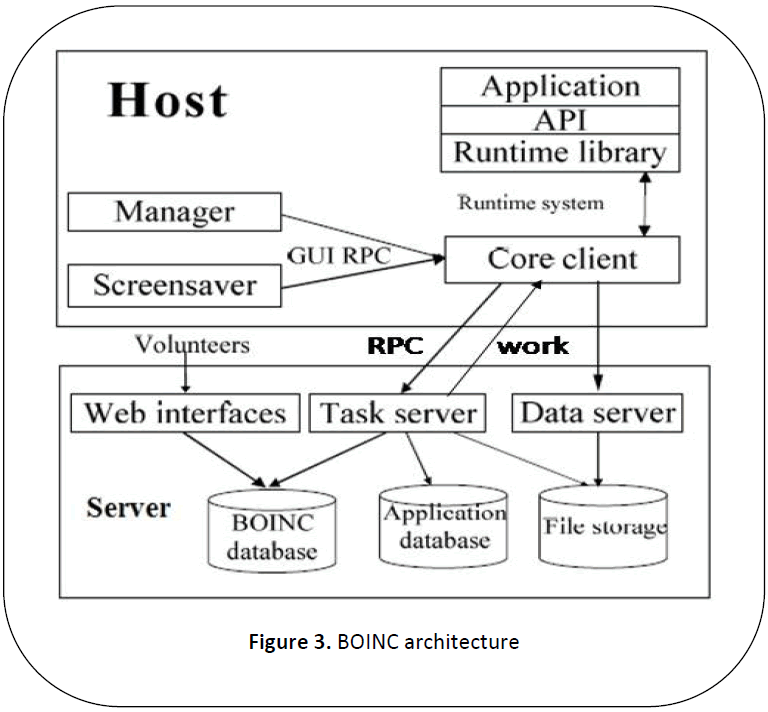
As explained in [13] BOINC has a very
scalable architecture which contains two
independent components, server software
component and client software component of
the middleware. Each client downloads an
application and part of the data to be
processed. It computes the results for the data
and sends the results back to server. Server
consolidates these results and returns the
solution to the user. The general architecture
of BOINC is as shown in Figure 3.
Figure 3: BOINC architecture
Server middleware software uses set
of daemons acting as various components to
perform the various tasks of sever as shown in Figure 3. Task server issues work to the client
and handle reports for completed tasks and
also handle the remote procedure calls
requested by the clients. Data server validates
the upload and access of files. It also checks
for the legitimacy of the files. There are many
other daemons not shown in figure such as
transitioner, validate, assimilater, file delete
that assists the server in completion of its
tasks. Any system that downloads the BOINC client middleware and connects to the project
by registering on their page can contribute
resources to those projects. The web pages of
the projects enable clients to select how their
resources can be used.
The main goal of BOINC is to
increase number of applications and increase
the participation of clients in volunteer
computing and retaining the participating
clients [19]. To achieve this goal, most of the
barriers to entry to public participation have
been taken care. In this section, few of the
barriers have been highlighted and how they
are solved in BOINC has been described.
Dealing with malicious and different
client architectures: Different client may send
back different results for the same data either
intentionally or unintentionally. Unintentionally
values may differ because of
difference in machine architecture, operating
system, compiler, and compiler flags. And
these values may vary more for floating point
numbers. Intentionally varying results are
primarily because of malicious clients that try
to break server application. Whether they are
intentional or intentional they are being taken
care using redundant computing where each
task is executed by X clients and those results
are checked to get a canonical result that’s
returned by at least Y number of clients. The
positive side affect of redundant computing is
that it is not affected by the failure of client
nodes.
User management
Success of applications in volunteer
computing depends on large participation of
user machines. Various methods have been
employed to increase, such as inventions of
3G Bridge to make BOINC compatible with
EDGeS [9] as explained in [20] and increasing
awareness about BOINC will have a positive
impact in participation as explained in [8]. The
other main challenge in user management is
retaining the participants as a negative review
by one participant will lead to loosing many participants and most of the participants
expect something in return. Hence BOINC
has come up with gaming technique to
introduce leader boards to keep participants
motivated in scientific applications. It also
uses customize web pages to socialize and
form teams for retaining most of the
participants.
Client mechanisms
Most of the participants are just happy
to download application version and work
unit from the server and execute the
application on that work unit and return back
the result. But few participants may be too
conservative to have some restrictions like
they will allow applications compiled by
themselves, or have platforms not supported
by the project. Even these clients can
participate in BOINC using anonymous
platform mechanism by downloading and
compiling the source of application and
informing server to send only workload to be
operated.
Server might have been using any
scheduling algorithm, but client knows the
best fit to optimize the resources. Hence
BOINC uses local scheduling policy where
the client can schedule operations such as
when it can receive projects and execute them
without compromising the performance of
client side applications.
Velvet assembler
Velvet [4] is a set of algorithms
implemented for de-novo assembly for short
reads. Each node of the de Bruijn graph
contains a sequence of overlapping k-mers.
Adjacent k-mers overlap by k - 1nucleotides.
Nodes are connected by directed edge where
the origin node overlaps with the destination
node. The nucleotide can be obtained by
traversing the graph given the first node and
the path information. The graph is constructed
and then errors if any removed as explained in
the following paragraphs.
The graph is created by first hashing
the reads for a given length and then searches
for the over lapping information among reads
to create a roadmap file. This roadmap file
contains either roadmap lines or non road
map lines. Roadmap lines specify the read
number and the non roadmap lines contain the
overlapping information about various reads.
It represents the information in 4 columns [12].
As an example, if the second reporting line
has four entries – ’1 25 0 5′ with k=21. It
means ROADMAP 2 has a 21-mer overlap
with ROADMAP 1 (first column). The
overlap starts from coordinate 25 of
ROADMAP 2 (second column) and
coordinate 0 of ROADMAP 1 (third column).
The overlap is 5 nucleotides or rather 5 kmers
long (fourth column). The
corresponding graph can be simplified my
merging adjacent nodes that are
uninterrupted.
Few errors usually nodes with less
coverage such as tips (at the ends of the
reads), bubbles (read errors or to nearby tips
connecting, and erroneous connections due to
cloning errors or to distant merging tips) are
to be removed. These errors are removed in
sequence starting with tips. Tips are errors at
the edge of the reads and it may be genuine
sequence. To eliminate only tips that are
erroneous conditions they use 2 measures:
length and minority count. Length help to
identify with genuine sequence with tips and
minority count helps in keeping the graph
away from complete erosion. Graph is
simplified again after removing tips.
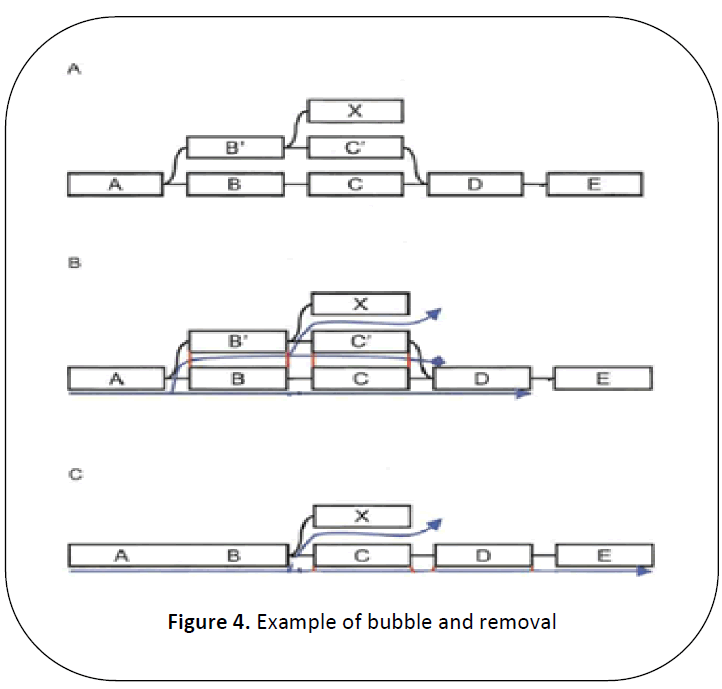
The bubbles are caused because of the
redundant paths that start and end at the same
node and contain similar sequences. The
redundant path is detected through a Dijkstralike
breadth-first search. The scan starts at a
node and traverse to other node till it scan a
node that is previous visited. Then it
backtracks to find the closest common
ancestor, and the sequence from both paths is
aligned to check if they are similar enough. If they are found to be similar, then those two
paths are merged by merging the longer path
into shorter path. The other nodes are linking
to the merged.
In Figure 4 nodes BC and B’C’ forms
a bubble, which can be detected because of
the previously visiting node in the second
traversal. The nodes are traced back to A as it
is the common ancestor. The sequences from
BC and B’C’ are extracted and found to be
similar enough. Hence the longer B’C’ is
merged with BC and the external node X is
connected by an edge from B.
Figure 4: Example of bubble and removal
After the graph is free from bubbles
and tips, other erroneous conditions are
removed from the graph using minimum
coverage. Since it is run after tour bus, it
doesn’t eliminate the unique connections
present in the graph.
Although there are many de novo
assembly algorithms, Velvet is chosen
because it generated accurate measured using
50 % median values called N50 lengths, its
Depth of Coverage at N50 length Plateau was
relatively low, and it required acceptable
computational resources [24]. Consequently, it is
feasible to use velvet for assembling large
sequences, such as those obtained for humans
and other mammals. Velvet uses larger
memory when compared with other tools to
store intermediate results. But this is not a
huge hindrance as memory cost has reduced a
lot.
Moving legacy applications to BOINC
As discussed earlier de novo assembly
requires huge memory and computational
requirements. Instead of acquiring a super
computer to enable de novo assembly, this
can be execute by gathering resources by
establishing a grid using BOINC as
middleware that gives the system architecture
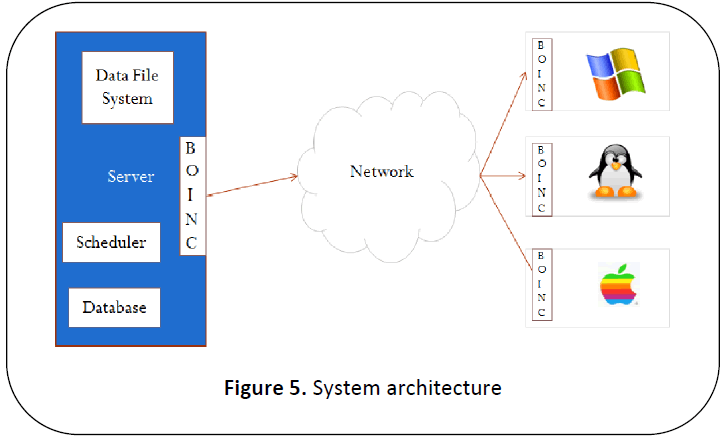
as in Figure 5.
Figure 5: System architecture
In the laboratory, any one of the
systems with sufficient resources usually
higher RAM and harddisk size capacity compared to the client machines is made as
the server or master as shown in Figure 5. This
system deals with co-ordination with clients.
BOINC server middleware software is to be
installed on this machine. The easiest way to
do so is to install middleware as part of virtual
image downloaded from BOINC website [11] on
to the server machine. The client machines
will have BOINC client software installed on
it. Server may also have additional scheduler
and database software. The data is stored
usually as File system either internal or on
Network Attached Storage (NAS) box.
The legacy applications such as velvet
described in previous section cannot be
directly used in this BOINC setup. These
applications are divided into two types to
enable their transfer to BOINC, the normal
applications that do not use graphics and the
set of graphical applications that has graphical
screen saver or some graphical interface.
There are two ways to move legacy
applications to BOINC. One method uses
wrappers and the other uses code changes.
For many of the existing applications
there is no source code available and they are
immutable. To use this kind of applications
with volunteer computing, BOINC provides a
wrapper which handles the communication
with the Core Client and executes the legacy
application as a sub process. It is configurable
but is not flexible enough to use for many
applications. It is implemented such that it
reads an xml (jobs.xml) that specifies the
execution sequence of the applications. To
overcome this setback a generic wrapper is
developed. This generic wrapper also called
as Gen Wrapper [3] provides a generic solution
to execute legacy applications. It utilizes a
shell scripting environment similar to POSIX
to control execution of applications and
processing of work unit. Gen Wrapper usually
consists of a Gitbox and Launcher
executables and a profile script to perform
platform specific preparations. Work unit,
referred to the set of data downloaded by a client, contains input files and a shell script
which should be platform independent.
Various versions of the same application can
be maintained at the server and is selected
using an xml file for the specific platforms.
If you are implanting a new
application or you have source code available
for existing applications you can use API
functions provided by BOINC to enable those
applications executable on grid you created.
These provide better check pointing in the
applications and can be optimized compare to
using wrappers. To use these applications we
need to add calls to BOINC initialization and
finalization routines. Convert all file open
calls to use physical file names by resolving
them from logical names. The code is then
linked with runtime libraries provided by
BOINC. These API are implemented in C++
and can be used with C usually. It has binding
that enable it to use with other languages like
FORTAN, JAVA and LISP.
Velvet in BOINC
Velvet has four stages: hashing the
reads into k-mers, constructing the graph,
correcting errors, and resolving repeats. Once
these stages are completed, they can be read
to get maximum contings. There are two
applications that perform these tasks: Velveth
and Velvetg. Velveth requires more resources
to construct graph and this graph is used by
Velvetg to give out the contigs that satisfy
given length and coverage details. Velveth is
modified to be used with BOINC such that
construction of graph is faster. We have
modified velveth to work with both windows
and different kinds of Linux implementations.
Since Velvet is an open source and is
implemented for Linux, we have used
BOINC API to modify certain sections of
code to be used with Linux clients. For
windows based clients we used to wrapper
based method with basic wrapper provided by
BOINC. The client is modified such that it
accepts and registers only velvet as and the default application to be executed using core
clients of BOINC.
Conclusions & Future Work
In this paper, explanation of how
scientific applications that require high
computation and storage requirements such as
de novo assembly can be solved without
using expensive super computers has been
discussed in detail. The application uses
resources that are idle from a cluster of
computers. The part of the application is
executed on part of the partial work unit
received from the server. The results are sent
back to the server for the execution of
remaining part. The solution is free from any
failures to clients.
We have implemented an existing de
novo assembly process using an existing
algorithm. In future, a new algorithm can be
designed and implemented from scratch to
optimize the use of resource as in
SETI@home and other projects currently in
BOINC. Once the gird is established and if
there is requirement for more resources then,
the same grid can be made public and
volunteers could be collected from all over
the world and acquire largest computing
power.
References
- Adam L. Beberg et al, “Folding@home: Lessons from Eight Years of Volunteer Distributed Computing”, IEEE 2009.
- An introduction to next generation Sequencing technology, white paper, https://www.illumina.com/technology/seq uencing_technology.ilmn.
- Attila Csaba Marosi et al, “GenWrapper: A Generic Wrapper for Running Legacy Applications on Desktop Grids”, IEEE 2009.
- Daniel R. Zerbino, Ewan Birney,” Velvet: Algorithms for de novo short read assembly using de Bruijn graphs”, Genome Res. 2008 18: 821-829. doi:10.1101/gr.074492.107
- David P. Anderson et al, “SETI@home”, communications of the ACM, 2002, 56-61.
- David P. Anderson, “BOINC: A System for Public-Resource Computing and Storage”, Proceedings of the Fifth IEEE/ACM International Workshop on Grid Computing, 2004, 1550-5510/04.
- David Stainforth et al, “Security Principles for Public-Resource Modeling Research”.
- David Toth et al, “Increasing Participation in Volunteer Computing”, IEEE International Parallel & Distributed Processing Symposium, 2011, 1878-1882.
- E. Urbach et al, "EDGeS: Bridging EGEE to BOINC and Xtrem Web, Journal of Grid Computing", 7(3) (2009), 335 -354.
- ERIC S LANDER, MICHAEs. L WATERMAN, “Genomic Mapping by Fingerprinting Random Clones: A Mathematical Analysis”, GENOMICS 2, 1988, 231-239.
- https://boinc.berkeley.edu/trac/wiki/VmS erver.
- https://www.homolog.us/blogs/blog/2011 /12/06/format-of-velvet-roadmap-file/.
- Huiping Yao et al, “Using BOINC Desktop Grid for High Performance Memory Detection”, IEEE, 2009, 1159- 1162.
- Jason R. Miller et al, “Assembly Algorithms for Next-Generation Sequencing Data”, Genomics. 2010 June, doi: 10.1016.
- Jennifer M. Schopf et al, “Monitoring the grid with the Globus Toolkit MDS4”, Journal of Physics: Conference Series 46 (2006) 521–525, doi: 10.1088/ 1742-6596/46/1/072.
- Lilian T. C. Franca et al, “A review of DNA sequencing techniques”, Quarterly Reviews of Biophysics 35, 2 (2002), pp. 169–200, DOI: 10.1017.
- Micheal J Litzkow et al, “Condor- a Hunter of idle Workstations”, 1987.
- Pavel A. Pevzner et al, “An Eulerian path approach to DNA fragment assembly”, PNAS, 2001, 9748–9753.
- Peter Darch, Annamaria Carusi|,” Retaining volunteers in volunteer computing projects”, The Royal Society, 2010, 4177-4192.
- Peter Kacsuk, “How to make BOINCbased desktop grids even more popular?”, IEEE International Parallel & Distributed Processing Symposium. 2011,1871-1877.
- Travis Desell et al, “Asynchronous Genetic Search for Scientific Modeling on Large-Scale Heterogeneous Environments”, IEEE, 2008.
- Xinhua Lin et al, “Recent Advances in CFD Grid Application Platform”, Proceedings of the 2004 IEEE International Conference on Services Computing, 2004.
- Yiming He et al, “De Novo Assembly Methods for Next Generation Sequencing Data”, Tsinghua Science and Technology, 2011, 500-514.
- Yong Lin et al, “Comparative Studies of de novo Assembly Tools for Next generation Sequencing Technologies”, Bioinformatics Advance Access, 2011.
- Zhenyu Li et al, “Comparison of the two major classes of assembly algorithms: overlap^layout^consensus and de-bruijngraph”, Briefings in Functional Genomics, 2011, 25-37.