Hiroshi Yasue1*, koji Doi1 and
Abdelfatteh El Omri2
1Department of Genetics, Tsukuba Gene Technology Laboratories, Ibaraki, Japan
2Department of Laboratory Medicine and
Pathology, Hamad Medical Corporation,
Doha, Qatar
- Corresponding Author:
- Yasue H
Department of Genetics, Tsukuba Gene
Technology Laboratories, Ibaraki, Japan
E-mail: hyasue@tsukuba-genetech.com
Received: January 11, 2021; Accepted: January 25, 2021; Published: February 1, 2021
Citation: Yasue H, Doi K, El Omri A (2021) Current Status of Gene Expression Analysis.
J Vet Med Surg Vol.5 No.1: 32.
Description
The change of biological state such as that of phenotypes in
organisms is definitely based on the change of gene expression.
The gene expression is the only path from genes to phenotypes.
Therefore, in order to understand the biological process change
in different organisms, gene expression analysis has been done
using various methods.
Alwine et al. from Stanford University described for the first time
the use of Northern Blot technique [1]. Few years later (from
1980), this method was frequently used. Northern Blot is based
on RNA electrophoresis in agarose gel and the transfer of the RNA
in the gel to nitrocellulose membrane, followed by hybridization
with a labeled probe specific to a given gene sequence [2].
However, several days are necessary to obtain results of the
Northern blot analysis for a single gene.
In 1984, the discovery of Polymerase chain reaction (PCR) by Kary
Mullis revolutionize science and let to the invention of Real-time
PCR [3]. This new technique has been applied through reverse
transcription of mRNA, showing exact copy number of mRNA
in a sample, though the number of genes for analysis in one
sample tube is limited up to four. Almost in parallel with usage
of the real-time PCR, microarray analysis has come to be used
for the expression analysis of huge number of genes at one
time. Microarray technique was introduced late 1977, then was
robotized by 1980-1990 to generate reproducible and repeatable
results, and it is only by 1995 that the technique was miniaturized
and the first publication using the word “microarray” was
published [4]. Microarray is based on the hybridization of probes
fixed on glass, each of which is specific to the corresponding
genes. These analyses are based on nucleic acids hybridization,
so that copy number controls are necessary for the calculation
of number of the targeted mRNA in the samples. In other words,
the direct comparison of real-time PCR values/signal intensity
of microarray analysis between samples doesn’t provide correct
results. The comparison should be done through the copy number
controls. In addition, these provide less information about slice
variants because of the following reasons; the primer pairs for
the real-time PCR/probe sequences for the microarray are usually
designed for limited exons of the target genes. In addition, mRNA
molecules are not analyzed as continuous RNA strand, so that,
it is not possible to constitute full-length transcripts, which
are particularly important information for the change of gene
expressions.
By the mid-2000s, RNA sequencing and its analysis tools such as
GeneStar have been available [5-7]. The gene expression studies
using the RNA sequencing are increasingly published. As is seen in
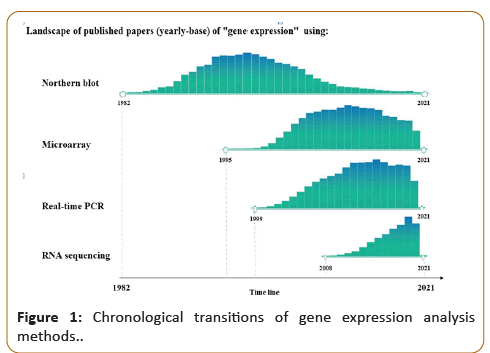
Figure 1, the chronological transition of gene expression analysis tools is represented by the number of publications over time.
Publication on Northern blot analysis started around 1982, and
showed its peak usage in 1998, and then started to decrease until
now. The Microarray and real time-PCR usage peaked around
2011 and 2015, respectively. Since the past few years, RNA
sequencing is increasingly used until now, though 2020 showed
a slight decrease in its usage. This can be partially attributed to
coronavirus pandemic that led to shut-down of universities and
research institutes all over the world, and the perturbation of the
global supply-chain logistics of genomic reagents and supplies.
Figure 1: Chronological transitions of gene expression analysis
methods.
RNA sequencing and the following in silico analysis can elucidate
not only the amounts of gene expressions but also the change
of gene splicing (change of exon usage). In addition, the digital
data generated by RNA sequencing would be used for direct
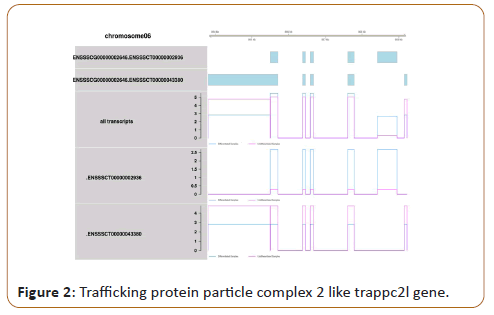
comparison between samples and even between experiments performed in different occasions. As an example, Figure 2 (not yet
published data) illustrated splicing variants and expression level
of exons of “Trafficking Protein Particle Complex 2 like TRAPPC2L”
obtained from RNA sequencing. This gene showed no significant
difference based on p-value (0.01<p<0.05) between samples,
but when investigated at respective exons, it showed significant
difference (p<0.01).
Figure 2: Trafficking protein particle complex 2 like trappc2l gene.
Discussion and Conclusion
This would provide deep insight for control of gene expression
leading to phenotypic difference. This kind of observation and
analysis cannot be obtained except by RNA sequencing, indicating
that RNA sequencing is one of promising tools to understand the
biological process of organisms deeply.
References
- Alwine JC, Kemp DJ, Stark GR (1977) Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci USA 74:5350-5354.
- Surzycki S (2000) Northern Transfer and Hybridization. Basic Tech Mole Biol 11:263-297.
- Deepak S, Kottapalli K, Rakwal R, Oros G, Rangappa KS, et al. (2007) Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr Genomics 8:234-251.
- Schena M, Shalon D, Davis RW, Brown PO. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-70.
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, et al. (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523-536.
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, et al. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 320:1344-1349.
- Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621-628.