Keywords
SARS-CoV-2; COVİD-19; Coronavirus; Receptor binding proteins;
Pandemic
Introduction
Background and Origin
Coronaviruses are a diverse group of viruses that cause infection
in animals and humans and they comprise a positive strand
genomic RNA. The members of this family have distinctive
morphological features and cause mild to severe respiratory
infections in the host body. However, the twenty-first century
began with the emergence of two pathogenic coronaviruses
known as severe acture respiratory syndrome (SARS-CoV) and
Middle East respiratory syndrome coronavirus (MERS/CoV) and
recent emergence of novel SARS-CoV-2 that were detected to
have had zoonotic origin (that are transmitted from animals to
humans) [1-3]. SARS-CoV-2, later known as coronavirus 2019
(COVID-19) was first detected in the city of Wuhan, China [4]
that cause mild to severe viral pneumonia followed by fever,
severe breathing problems (dyspnea), chest discomfort and lung
infiltration [4]. İn comparison with MERS-CoV and SRAS-CoV,
SARs-CoV-2 spreads faster and wider and so far has caused a
remarkable threat to human race worldwide [5]. The outbreak
was reported to have originated from seafood market in Wuhan,
China that sells live animals and poultry. Although the first cases of COVID-19 are dated back to late December 2019, the first
reported case was reported on 8th of December 2019 [6].
RNA sequencing analysis of the isolated virus from infected
patients detected novel SARS-CoV-2 to be belonged to
β-coronovarisu subgenus of the Coronoviridae family which is
part of virus order known as nidovirales [7-11] with zoonotic
nature of transmission. Further nosocomial infections at hospitals
and family members identified the transmission to have occurred
from human to human [12-14].
In the statement made by the world health organization on
January 31, 2020, Public Health Emergency of International
Concern (PHEIC) listed COVİD-19 as a posing a threat for to many
countries and called for international precaution. The most common features of COVID-19 in patients are respiratory tract
infection which are detected with computer tomography (CT) and
chest X-ray in the acute period [15]. In a mild case of COVID-19,
patients may experience mild respiratory infections which can
turn to become acute with the severe case of pneumonia. Based
on WHO annuncement, Approximately 80% of patients were
asymptomatic or developed mild symptoms, while 20% are
detected with serious clinical conditions which are particularly
observed with older people or the individuals with hypertension,
cardiovascular diseases, chronic obstructive pulmonary disease,
diabetes, obesity, and any malignancy. Genetic analysis of viral
isolates from infected patients showed SARS-CoV-2 nucleotides
to have nearly 88% similarity with bat-like SARS coronavirus
indicating a zoonotic nature of the virus. Bats were found to
be the reservoir hosts for SARS-CoV-2 [16]. The virus is thought
to be transmitted from person to person by droplets that can
spread from the infected people with talking, sneezing/coughing,
touching the mouth, nose, or eye mucosa after the contact with
infected areas. The virus has a mean incubation period of 6.4 days
[17]. Clinical manifestations in COVİD-19 patients, including fever,
cough, and tiredness are similar to that of SARS-CoV and MERSCoV
cases. Shortness of breath, chills, sore throat, loss of taste or
smell and chest pain are listed among other disease symptoms
[15]. The severity of the disease varies with the immune system
response as well as pre-existing clinical conditions from person
to person. As the current treatments have been insufficiently
effective on the patients, the scientists and researchers have
been trying to develop the efficient therapy with minimal shortand
long-term side effects and to reduce the mortality rate
worldwide.
Phylogenetic analysis of viral isolates from the patients reveals
various evolutionary processes of the virus. Epidemiological studies have shown that clinical outcomes are greatly influenced
by the individual's age, gender, complications, and other possible
unknown clinical parameters [18]. All societies should take the
necessary precautions tackle the current pandemic.
The origin and structural features of SARS-CoV-2
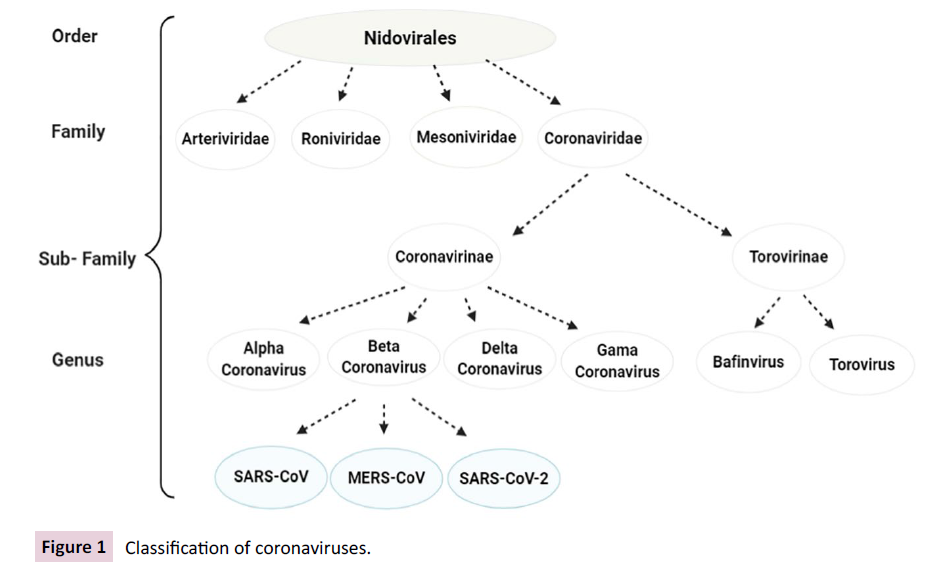
Coronaviridae virus families constitute the Nidovirales
Arteriviridae, Roniviridae, and Mesoniviridae (Figure 1). SARSCoV-
2, along with SARS-CoV and MERS-CoV, is a member of the
subfamily Coronavirinae called Beta-coronavirus [19]. Four other
types of disease-causing CoVs are endemic in humans: 229E,
OC43, NL63 and HKU and they cause common cold symptoms.
SARS-CoV causes serious respiratory distress and death in
infected cases. The first SARS-CoV case was identified in 2003.
The Middle East respiratory syndrome coronavirus (MERS-CoV)
was described highly pathogenic in 2012. SARS-CoV-2 is the 7th known type of coronavirus that cause severe symptoms after
identification of MERS and SARS, while HKU1, NL63, OC43, and
229E have been associated with milder symptoms [20].
Figure 1: Classification of coronaviruses.
Coronaviruses are the largest known RNA viruses with no
segments, single-stranded, positive chain and genome’ length
ranges from 26 to 32kb [21,22]. The typical coronavirus genome
organization is 5′-1a-1b-S-E-M-N-3′, and this sequence is common
among coronaviruses. The partially overlapping open frames 1a
and 1b makeup two-thirds of the viral genome and encode all the
elements necessary for viral RNA synthesis. This region is defined
as the replicase gene [23]. Two large polyprotein (pp1a and pp1b)
are encoded from coronavirus replicase genes (ORF 1a and ORF
1b). Structural proteins of the coronavirus are encoded by almost
a third of the viral genome [23].
SARS-Cov-2 includes nonsegmented, single-stranded, positivesense
genomic RNA, with 5 'cap structure and 3' poly-A tail similar to other coronaviruses [24]. The genome sequence of the SARSCoV-
2 BatCoV has 96.2% similarity with RaTG13 coronavirus in
bats and 79.5% similarity with SARS-CoV [25]. Thus, bat is still
considered the most likely species for the 2019 new coronavirus
(SARS-CoV-2) source.
Generally, coronaviruses encode four structural proteins in their
genomes [26]. Genomic RNA of the coronaviruses is packaged by
the viral nucleocapsid (N) protein. Three other structural proteins
of coronavirus are Spike (S), Membrane (M), and Envelope (E)
proteins that form the envelope of virions. These four structural
proteins are common among coronaviruses; (N, M, S, E). The S
glycoprotein provides the coronaviruses' entry into the host
cell, a class I virus fusion protein [27]. Coronavirus membrane
(M) protein is the most common structural protein and plays a
vital role in forming virus particles that interacts with N protein
in many coronavirus species. Coronavirus nucleocapsid (N)
protein is required for the efficient packaging of viral genomic
RNA into the helical ribonucleoprotein (RNP) complex during the
formation of virions [28]. Coronavirus envelope (E) protein is the
main component of the viral envelope. It also plays an important
role in releasing virions from the cell and the regulation of other
cellular functions [29]. The S genome of the SARS-CoV-2 shows
93.1% high similarity with the RaTG13 S genome, while it has a
similarity rate of less than 75% with SARS-CoV [25]. It is stated
that the highest similarity between SARS-CoV-2 and SARS-CoV is
in the N terminus [30,31].
Coronaviruses are introduced into the cell by the S protein of
the virus. While the mouse coronavirus enters the MHV cell via
the CEACAM1 receptor, SARS-CoV can enter the cell by binding
S protein to angiotensin-converting enzyme 2 (ACE2). When the
S1 subunit of the S protein is attached to the host cell receptor,
the S protein undergoes a series of structural (conformational)
changes to ensure fusion with the host cell membrane [21]. The
SARS-CoV-2-S protein binds ACE2 with higher affinity than
SARS-CoV Receptor Binding Domain (RBD) [32]. After S protein
binds with the ACE2 receptor on the host cell, the S protein is
cleaved by host cell proteases and viral-host membrane fusion
occurs. Depending on the type of virus, it the fusion may occur
in the host cell's plasma membrane or acidified endosomes
after receptor-mediated endocytosis. The binding of the
virus with host cell receptors is a critical factor for the virus's
pathogenesis [33,34]. A better understanding of receptor binding
and protease effect relative characteristics will also help predict
how the coronaviruses infect humans and adapt to the host cell.
Diagnosis of COVID-19 and current treatment
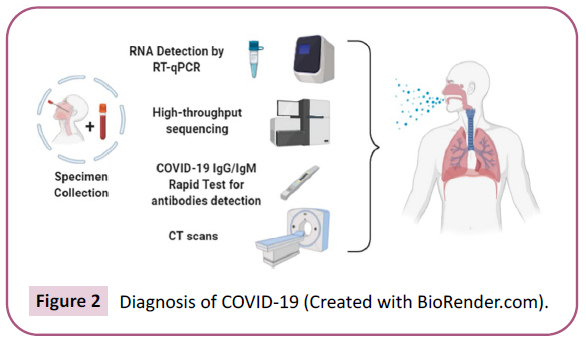
Epidemiological history, clinical findings, nucleic acid detection,
and CT scan are mainly used to diagnose COVID-19 (Figure 2).
Additionally, the IgM/IgG Fluorescence Immunochromatographic
Assay method (POCT) produces rapid early diagnosis results. In
cases with strong epidemiological relationship with COVID-19 infection,
the application of serological tests in serum samples taken
in the acute or convalescent phase may support the diagnosis.
For this purpose, enzyme-linked immunosorbent assay (ELISA)
or rapid antibody tests that detect IgM/IgG help investigate the
ongoing outbreak and provide a retrospective evaluation of the attack rate and the severity of the epidemic. İn addition, recent
studies on viral isolates collected from stool samples show SARSCoV-
2 replication in a stool sample [35] that could be considered
as a potential source of viral isolation.
Figure 2: Diagnosis of COVID-19 (Created with BioRender.com).
The clinical symptoms of patients infected with SARS-CoV-2 are
different from each other, that is, atypical. In cases, fever, shortness
of breath and radiological findings are compatible with bilateral
pulmonary infiltrate. Although different protocols were published
to target N, E and S genes for diagnosis of COVİD-19, it is
more efficient to adopt more accurate technique such as RT-qPCR
can identify SARS-CoV-2 virus based on N gene and RdRP, ORF1a
and reduce the errors in the test result [36].
Among drugs that are candidates for treating COVID-19, reevaluation
of old medications for antiviral therapy is a usual
strategy because their safety profile, side effects, posology, and
drug interactions are well known [37,38]. Apart from antiviral
treatment, vaccine development studies, efficacy studies of
biological agents (MABs), and different antiparasitic drugs are
being tested. In the absence of the vaccine, an international
investigation "Solidarity" was launched by WHO to overcome the
pandemic. The drugs included in this study are listed as lopinavir
and ritonavir, lopinavir and ritonavir interferon beta, chloroquine
and remdesivir [39].
Interleukin-6 (IL-6) pathway inhibitors are another alternative form
of treatment. Tocilizumab was developed against recombinant
human IL-6 receptor monoclonal antibody. In COVID-19 patients,
high IL-6, IL-1 and TNF-α levels were observed with severe clinical
manifestations that result in "cytokine storm". Tocilizumab is an
immunosuppressive drug against the IL-6 receptor in COVID-19
patients who developmed severe pulmonary symptoms. Although
the use of Tocilizumab against IL-6 receptor in the cytokine storm
is supported, there is no published clinical data with proven
effectiveness [40].
Kinases are another alternative way of therapy. p21-activated
protein kinases (PAKs) are small (p21) GTPases that contain
members of the Cdc42 and Rac families. It has been shown
that PAK1 has an important role in the entry, replication and
spread of many important viruses, including influenza and HIV
[41,42]. Coronaviruses use macropinocytosis to enter the host
cell, and this process has been shown to depend on PAK1 activity.
Thus, this suggests that PAK1 inhibitors can be valuable in the
treatment of COVID-19 [43].
Vitamin C improves endothelial functions, reduce blood pressure
and risk of atrial fibrillation, and decrease contrast-induced kidney
damage. Studies indicate vitamin C as an alternative supportive
treatment method against hyperinflammation caused by viral
infection [44-46]. Many studies suggest anti-inflmmatory effect
of IV vitamin C that blocks release of IL-6 in the endothelial cells
that are induced by endothelin-1 (ET-1) [47-52]. Although some
papers propose the effectiveness of IV application of vitamin C on
severe case of COVID-19 by suppressing the onset of COVID-19
penumonia, further clinical studies are required to confirm
vitamin C as a treatment mean to suppress severe pulmonary
symtoms caused by SARS-CoV-2.
Pathogenesis and Immunity of SARS-COV-2
As it is known, ARDS (Acute Respiratory Distress Syndrome)
is pulmonary edema caused by increased permeability of
the capillaries in the lungs. It causes the fluid transfer to the
pulmonary region as a result of acute inflammatory lung injury.
Here, the causes damaging the alveolocapillary membrane may
be due to the direct airway, such as pneumonia, aspiration, or
indirect hematogenous causes such as sepsis and uremia. In the
study by Xu et al., [53] ARDS was shown to be one of the main
causes of death in severe COVID-19.
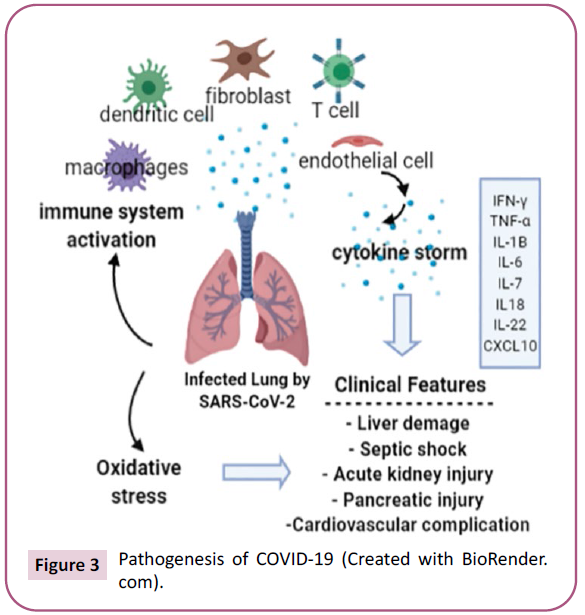
Cytokine storm is one of the primary mechanisms for ARDS
formation. It is an over-developing systemic inflammatory
response from uncontrolled release of pro-inflammatory
cytokines (IFN-a, IFN-y, IL-1, IL-6, IL-12, IL-18, IL-33, TNF-α) and
chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10) by immune
effector cells in SARS-CoV infection. This uncontrolled immune
response was stated to be closely related to the determination of
COVİD-19 severity. Also, increased interleukin-6 (IL-6) levels with
“cytokine storm” have been observed in COVID-19 patients with
severe clinical findings [54] (Figure 3). In the cytokine storm seen
in severe COVID-19 patients, an immunosuppressive drug against
was recommended against the IL-6 receptor, targeting the IL-6
pathway [55].
Figure 3: Pathogenesis of COVID-19 (Created with BioRender.com).
MAS (macrophage activation syndrome) may also develop in
COVID-19 disease, but there is no evidence on the frequency of
occurrence and definitive treatment. It is believed that 10% of
COVİD-19 patients may be in the critical condition and cytokine
storm caused by MAS may worsen the prognosis. For this reason,
the correct and timely development of effective treatment
against the cytokine storming can efficiently reduce the mortality
rate. The case series and observations reported to date indicate
that the MAS findings in COVID-19 patients generally show
similarity to MAS tables in hereditary HLH (hemophagocytic
lymphohistiocytosis) rheumatic diseases [56]. However, MAS/
HLH findings may not develop in all patients depending on the
disease's course. The scores or criteria used in the diagnosis of
other conditions may not always help decide the best treatment
method [56].
The presence of signs such as persistent fever, an elevated level
of C-reactive protein (CRP) and ferritin values, a high D-dimer,
lymphopenia and thrombocytopenia, liver deterioration function
tests, hypofibrinogenemia or increased triglyceride values may
indicate that the MAS deteriorate the patients’ conditions. MAS is a complication that requires close follow-up and early treatment.
When the patient diagnosed, it can become much more difficult
or impossible to suppress the developing cytokine storm within
hours [56].
Possible impact of frequent mutations in the
novel SARS-CoV-2 genome on pathogenicity and
transmission ability
The increasing epidemiological and clinical evidence implicates
that the SARS-CoV-2 has stronger transmission power and
lower pathogenicity than SARS-CoV [57]. However, the high
transmission mechanism of SARS-CoV-2 has not been clarified
yet. Researches have found that the virus is rapidly mutating.
As a result of mutation, they have discovered three different
"variants" of COVID-19 consisting of closely related strains called
"A", "B" and "C". Based on SARS-CoV-2 evolutionary tree, and it
was stated what the mutations occurred during the viral spread
from Wuhan to Europe and North America. Thus, the scientists
have been investigating to determine the factors causing the
evolution of SARS-CoV-2 to type-A, type-B and type-C with limited
genome data [58].
The coronavirus subtype "A" variant is most closely associated
with the virus in both bats and pangolins. Researchers have
described as "the source of the epidemic". The type "B" is derived
from "A" by two mutations. Later, a type C variant formed which
derived from B type coronavirus with a uniform mutation. Type
A has been found in Americans who are living in Wuhan and
patients diagnosed in the United States and Australia. While type
B coronavirus is common throughout East Asia, the C variant is
predominantly in Europe; it has been expressed as the primary
type of virus found in patients from France, Italy, Sweden and the
UK [59].
Detection of mutations occurring in the SARS-CoV-2 genome is
important to understand the evolution of the virus and reveal
genotype changes that occur during the transmission of SARSCoV-
2. Genotyping studies have demonstrated a few common
mutations in SARS-CoV-2 genomes. It has been stated that the
mutations might be related to the virus permeability and the
extent of pathogenicity. Detected mutations are found mostly
in S protein, RNA polymerase, RNA primase, and nucleoprotein,
which are essential for the vaccine efficacy [58]. For this reason,
mutation analysis might be the results that support the literature
ineffective treatment studies to prevent SARS-CoV-2 coronavirus
infection.
Discussion
A recent outbreak of COVID-19 has led to global and panic and
cost more than a million lives worldwide. Genome analysis of
SARS-CoV-2 remarked on the significance of spike (S) glycoprotein
that maintains viral invasion by binding to angiotensin-converting
Enzyme 2 (ACE2) [60,61]. SARS-CoV-2, similar to other RNA
viruses, display high mutation rate that may affect the virus's
pathogenicity [62] and consider the importance of S protein in
SARS-CoV-2 virulence, a mutation on the spike region is likely to
enhance the virus's pathogenicity [63]. S protein was shown to
undergo mutation owing to a receptor-binding domain (RBD)
of S protein, the most variable genomic component of SARSCoV-
2 [64]. Recent studies detected that infected patients with
the SARS-CoV-2 with substitution mutation of Asp to Gly in 614
position (D614G) in the spike glycoprotein show higher viral loads
in the upper respiratory tract [65-67] and this could suggest the
impact of the mutation on the transmission rate. Moreover,
the D614G strain also manifests stronger binding affinity to
angiotensin-converting (ACE2) [68,69] that could require further
investigation of the patients' pathogenicity and treatment. The ex
vivo and in vivo studies of D614G SARS-CoV-2 strain on different
cell lines detected higher replication rate of the strain in upper
respiratory tract cell lines with larger numbers of human ACE2
(hACE2) receptors [70,71]. However, further analysis of hamster
did not signify a higher pathogenicity rate with the studied strain
[4,70].
Further studies suggest a higher prevalence rate of D614G SARSCoV-
2 strain in younger ages which could relate to the higher
replication rate [72,73]. However, the follow-up examination
and observation could not confirm the effect of the mutation on
virulence or pathogenicity [72]. Similar to other viruses' features,
investigation on different types of SARS-CoV-2 shown to have a
higher transmission rate was followed by lower pathogenicity
degree [72-74]. Genomic analysis of SARS-CoV-2 on patients in
China detected several single nucleotide variants (SNVs) on S
protein that each manifested noticeable changes in the virus's
pathogenicity [75]. Furthermore, mutations on nonstructural
proteins (nsps) of SARS-CoV-2 was described to be the cause
virus's high contagion rate compared to that of SARS-CoV [76,77].
Some other studies on different mutation positions affect SARS-CoV-
2 pathogenicity and decrease virulence. Study on full-length
SARS-CoV-2 discovered 81 base pair deletion on AZ-AU2923
genome, an open reading frame (ORF)7a gene that synthesizes
the accessory proteins replication, infection, and proliferation of
the virus inside host body [66,78]. Detected substitution of Serine
by Arginine (S247R) in S1 subunit of S protein was in the SARSCoV-
2 isolates of some patients [35]. Although the position is
within the N-terminal domain of S1 subunit that is not directly part
of S subunit binding to ACE2 receptors, N-terminal is adjacent to
C-terminal that binds to ACE2 receptors. Mutation in this position
could affect the binding affinity of the virus. Confirmation of such
possibilities needs further investigation. İndicated substitution of
Asparagine by Arginine in 439th position (N439R) in RBD increase
SARS-CoV-2 binding affinity to ACE2 receptors and enhance viral
transmission from human to human [79,80]. However, detected
various mutation points on SARS-CoV-2 RBD, the mutation rate
on the virus reported to be remarkably low compared to other
members of coronavirus families. On the contrary, the higher viral
loads of SARS-CoV-2 in humans are likely to result in mutation at
the infection period. Even though almost all reported mutations
did not display increased pathogenicity of the virus, more than
one mutation in RBD is likely to enhance virulence. However,
further in vitro and in vivo investigations are required to confirm
a change in viral pathogenicity that can lead to drug resistance
or further treatments. Investigating these mutations in the SARSCoV-
2 genome whether it can affect the function of the structural
genes and the pathogenicity of SARS-CoV-2, revealed that the
mutation analysis is critical for the drug and vaccine development
against COVID-19 in the stage of the pandemic.
Conclusion
Coronaviruses have known as the common cause of death for
many years, and with the emergence of the new type of SARSCoV-
2 in early 2020, they pose a more severe threat to human
health, spread to the whole world in a short time, causing
thousands of cases and hundreds of deaths. Within the scope of
many molecular studies that are being conducted on different
populations on a global basis, it is thought that COVID-19 outbreak
analysis will also contribute to the control of the pandemic
in terms of providing epidemiological data, investigating viral
resistance development mechanisms, directing treatment and
clinical monitoring of the disease.
Authors Contributions
Şeyda Demirkol drafted the paper and prepared the images
and figures. Nasim Kherad collected the recent articles and
information on COVID-19 and drafted the paper. Elif Aslan helped
in data analysis and contributed in data collection. Saadet Busra
Aksoyer Sezgin, Sema Ketenci, and Ebru Nur Ay helped in writing
the paper and arranging the citation. Meryem Alagoz and Ilhan
Yaylım did the study design, checked the information and data
accuracy and finalized the draft.
References
- Cui J, Li F, Shi ZL (2019) Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 17: 181-192.
- Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, et al. (2020) The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Intl J Infect Dis 91: 264-266.
- Wu JT, Leung K, Leung GM (2020) Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet 395: 689-697.
- Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733.
- Deng SQ, Peng HJ (2020) Characteristics of and public health responses to the coronavirus disease 2019 outbreak in China. J Clin Med 9: 575.
- Wu Z, McGoogan JM (2020) Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. Jama 323: 1239-1242.
- Cong Y, Verlhac P, Reggiori F (2017) The interaction between nidovirales and autophagy components. Viruses 9: 182.
- Gorbalenya AE, Baker SC, Baric RS, De Groot RJ, Drosten C, et al. (2020) The species severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5: 536–544.
- Jiang S, Du L, Shi Z (2020) An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect 9: 275-277.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579: 265-269.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. BioRxiv.
- Chan JFW, Yuan S, Kok KH, To KKW, Chu H, et al. (2020) A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. The lancet 395: 514-523.
- Wang R, Zhang X, Irwin DM, Shen Y (2020) Emergence of SARS-like coronavirus poses new challenge in China. J Infect 80: 350-371.
- Wong SCY, Kwong RS, Wu TC, Chan JWM, Chu MY, et al. (2020) Risk of nosocomial transmission of coronavirus disease 2019: an experience in a general ward setting in Hong Kong. J Hosp Infect 105: 119-127.
- Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, et al. (2003) Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. The Lancet 362: 1353-1358.
- Hao P, Zhong W, Sonz S, Fan S, Li X (2020) Is SARS-CoV-2 originated from laboratory? A rebuttal to the claim of formation via laboratory recombination. Emerg Microb Infect 9: 545-547.
- Sheahan TP, Sims AC, Leist SR, Schäfe A, Won J, Brown AJ, et al. (2019) Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 11: 1-14.
- Lorenzo-Redondo R, Nam HH, Roberts SC, Simons LM, et al. (2020) A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EbioMedicine 62: 103112.
- Rehman SU, Shafique L, Ihsan, A, Liu Q (2020) Evolutionary trajectory for the emergence of novel coronavirus SARS‑CoV‑2. Pathogens 9: 240.
- da Costa VG, Moreli ML, Saivish MV (2020) The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch Virol 165: 1517-1526.
- Perlman S, Netland J (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7: 439-450.
- Rosado JA, Jenner S, Sage SO (2000) A role for the actin cytoskeleton in the initiation and maintenance of store-mediated calcium entry in human platelets: evidence for conformational coupling. J Biol Chem 275: 7527-7533.
- Siddell S, Wege H, Ter Meulen V (1983) The biology of coronaviruses. J Gen Virol 64: 761-776.
- Ashou HM, Elkhatib WF, Rahman M, Elshabrawy HA (2020) Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens 9: 186.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579: 270-273.
- Ziebuhr J (2004) Molecular biology of severe acute respiratory syndrome coronavirus. Current opinion in microbiology 7: 412-419.
- Bartlam M, Yang H, Rao Z (2005) Structural insights into SARS coronavirus proteins. Curr Opin Struct Biol 15: 664-672.
- Satija N, Lal SK (2007) The molecular biology of SARS coronavirus. Ann N Y Acad Sci 1102: 26.
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, et al. (2003) The genome sequence of the SARS-associated coronavirus. Science 300: 1399-1404.
- Kumar S, Maurya VK, Prasad AK, Bhatt ML, Saxena SK (2020) Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virusdisease 31: 13-21.
- Morse JS, Lalonde T, Xu S, Liu WR (2020) Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem 21: 730-738.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, et al. (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367: 1260-1263.
- Tufan A, Guler AA, Matucci-Cerinic M (2020) COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turk J Med Sci 50: 620-632.
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, et al. (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271-280.
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, et al. (2020) Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. MedRxiv.
- Kubina R, Dziedzic A (2020) Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics 10: 434.
- Colson P, Rolain JM, Raoult D (2020) Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents 55: 105923.
- Colson P, Rolain JM, Lagie JC, Brouqui P, Raoult D (2020) Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents 55: 105932.
- Uddin M, Mustafa F, Rizvi TA, Loney T, Suwaidi HA, Al-Marzouqi AHH, et al. (2020) SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses 12: 526.
- Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, et al. (2020) COVID-19: Consider cytokine storm syndromes and immunosuppression. The lancet 395: 1033-1034.
- Pascua PNQ, Lee JH, Song MS, Park SJ, Baek YH, et al. (2011) Role of the p21-activated kinases (PAKs) in influenza A virus replication. Biochem Biophys Res Commun 414: 569-574.
- Van den Broeke C, Radu M, Chernoff J, Favoreel HW (2010) An emerging role for p21-activated kinases (Paks) in viral infections. Trends Cell Biol 20: 160-169.
- Freeman MC, Peek CT, Becker MM, Smith EC, Denison MR (2014) Coronaviruses induce entry-independent, continuous macropinocytosis. Mbio 5.
- Ashor AW, Lara J, Mathers JC, Siervo M (2014) Effect of vitamin C on endothelial function in health and disease: a systematic review and meta-analysis of randomised controlled trials. Atherosclerosis 235: 9-20.
- Juraschek SP, Guallar E, Appel LJ, Miller III ER (2012) Effects of vitamin C supplementation on blood pressure: a meta-analysis of randomized controlled trials. Am J Clin Nutr 95: 1079-1088.
- Sadat U, Usman A, Gillard JH, Boyle JR (2013) Does ascorbic acid protect against contrast-induced acute kidney injury in patients undergoing coronary angiography: a systematic review with meta-analysis of randomized, controlled trials. J Am Coll 62: 2167-2175.
- Erol A (2020) High-dose intravenous vitamin C treatment for COVID-19.
- Fowler A, Jonathon D, Duncan R, Morris EP, DeWilde C (2019) Effect of Vitamin C Infusion on Organ Failure and Biomarkers of Inflammation and Vascular Injury in Patients With Sepsis and Severe Acute Respiratory Failure:The CITRIS-ALI Randomized Clinical Trial. JAMA 322: 1261-1270,
- Fujii T, Luethi N, Young PJ, Frei DR, Eastwood GM, et al. (2020) Effect of vitamin C, hydrocortisone, and thiamine vs hydrocortisone alone on time alive and free of vasopressor support among patients with septic shock: the VITAMINS randomized clinical trial. Jama 323: 423-431.
- Gao Y, Li T, Han M, Li X, Wu D, et al. (2020) Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID‐19. J Med Virol 92: 791-796.
- Liu B, Li M, Zhou Z, Guan X, Xiang Y (2020) Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)?. J Autoimmun 102452.
- Marik PE (2020) Vitamin C: an essential “stress hormone” during sepsis. J Thorac Dis 12: S84.
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, et al. (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 8: 420-422.
- Channappanavar R, Perlman S (2017) Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol 39: 529-539.
- Dzobo K, Chiririwa H, Dandara C, Dzobo W (2021) Coronavirus disease-2019 treatment strategies targeting interleukin-6 signaling and herbal medicine. Omics: J Integrat Bio 25: 13-22.
- Soy M, Atagunduz P, Atagunduz I Sucak GT (2020) Hemophagocytic lymphohistiocytosis: a review inspired by the COVID-19 pandemic. Rheumatol Int 1-12.
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, et al. (2020) Clinical characteristics of coronavirus disease 2019 in China. New Eng J Med 382: 1708-1720.
- Yin C (2020) Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics 112: 3588-3596.
- Forster P, Forster L, Renfrew C, Forster M (2020) Phylogenetic network analysis of SARS-CoV-2 genomes. Proceedings of the National Academy of Sciences 117: 9241-9243.
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, et al. (2020) Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci 6: 315e331.
- Yuan Y, Cao D, Zhang Y, Ma J, Qi J, et al. (2017) Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8: 1-9.
- Duffy S (2018) Why are RNA virus mutation rates so damn high?. PLoS biology 16: e3000003.
- Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94.
- Lv L, Li G, Chen J, Liang X, Li Y (2020) Comparative genomic analysis revealed specific mutation pattern between human coronavirus SARS-CoV-2 and Bat-SARSr-CoV RaTG13. BioRxiv.
- Korber B, Fischer WM, Gnanakar S, Yoon H, Theiler J, et al. (2020) Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182: 812-827.
- Korber B, Fischer W, Gnanakara SG, Yoon H, Theiler J, et al. (2020) Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. BioRxiv.
- Lorenzo-Redondo R, Nam HH, Roberts SC, Simons LM, Jennings LJ,et al. (2020) A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EBioMedicine 62: 103112.
- Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, et al. (2020) Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183: 739-751.
- Mansbach RA, Chakraborty S, Nguyen K, Montefiori DC, Korber B, et al. (2021) The SARS-CoV-2 spike variant D614G favors an open conformational state. Sci Adv 7: eabf3671.
- Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, eta l. (2020) SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370: 1464-1468.
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, et al. (2020) SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182: 429-446.
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, et al. (2021) Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell 184: 64-75.
- Walker PG, Whittaker C, Watson OJ, Baguelin M, Winskill P, et al. (2020) The impact of COVID-19 and strategies for mitigation and suppression in low-and middle-income countries. Science 369: 413-422.
- Young BE, Fong SW, Chan YH, Mak TM, Ang LW, et al. (2020) Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. The Lancet 396: 603-611.
- Yao HP, Lu X, Chen Q, Xu K, Chen Y, et al. (2020) Patient-derived mutations impact pathogenicity of SARS-CoV-2. Cell Disc 76.
- Angeletti S, Benvenuto D, Bianchi M, Giovanetti M, Pascarella S, et al. (2020) COVID‐2019: the role of the nsp2 and nsp3 in its pathogenesis. J Medi Virol 92: 584-588.
- Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, et al. (2004) Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78: 13600-13612.
- Holland LA, Kaelin EA, Maqsood R, Estifano B, Wu LI, et al. (2020) An 81-nucleotide deletion in SARS-CoV-2 ORF7a identified from sentinel surveillance in Arizona (January to March 2020). J Virol p. 94.
- Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiol 5: 562-569.
- Shang J, Ye G, Shi K, Wan Y, Luo C, et al. (2020) Structural basis for receptor recognition by the novel coronavirus from Wuhan. Nature 581: 221-224.