Research Article - (2017) Volume 3, Issue 2
Tamayo C1, Urzua U1, Díaz J1, Miranda VR2, Chahuan M3, Campos M3, Zambrano A14, Benitez H4, Marin P4 and Adonis M5*
2Hospital Barros Lucos Trudeau, Chile
3Hospital San Borja Arriaran, Chile
4Regional Hospital of Antofagasta, Chile
5Program of Cellular and Molecular Biology ICBM CeTeCancer, Avenida Independencia 1027, Santiago, Chile
*Corresponding Author:
Marta Adonis
Program of Cellular and Molecular Biology ICBM CeTeCancer
Avenida Independencia 1027, Santiago, Chile
Tel: 562994498609
E-mail: madonis@med.uchile.cl
Received date: June 16, 2017; Accepted date: July 18, 2017; Published date: July 24, 2017
Citation: Marta A, Carolina T, Ulises U, Lionel G, Jose D (2017) Circulating Free DNA in Serum and Genomic Alterations of Lung Cancer Patients and High Risk Individuals Exposed to Environmental Pollution. Biomark J. 3:12. doi: 10.21767/2472-1646.100033
Copyright: © 2017 Marta A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
The implementation of early-detection technologies and prognostic biomarkers are imperative, specifically in cities highly exposed to environmental carcinogens, such as PAHs and Arsenic. This study proposed to evaluate cfDNA levels and determine CNAs in serum cfDNA in LC and PNL patients. The study enrolled a total of 107 people, from the Metropolitan (N=51) and Antofagasta (N=56) regions, with medium or high LC risk. The cfDNA was isolated from serum aliquots using Qiagen QIAamp DNA mini kit. HMMs analysis in cfDNA, was used as a probabilistic model to determine an unknown sequence and defined Minimal Common Regions (MCRs) of recurring DNA gains and losses. This study shows that cfDNA levels increased during malignancy of LC but not significantly during premalignant lesions or pre-invasive stages. The cfDNA could be used as a complementary tool to prognosis and evolution of LC, with high precision (specificity of 87.3% and PNV of 76.4%). Additionally, the cfDNA carry some chromosomal aberrations related with the lung tumorigenesis process that could be used to confirm a suspected case of LC. Within these alterations our results suggest the loss of specific chromosomic fragments, contain genes related with inflammation response (SERPING1), humoral immune response (IGCK), innate immunity and epithelial integrity (CTNN1), tumor suppressor (MAP2K4, DMTF1 and ABCB4), and response to oxidative stress (COX10). In conclusion, these biomarkers in cfDNA, could have a great potential as screening methods in population with high risk for LC.
Keywords
Lung cancer; Early detection; cfDNA; CNVs; Biomarker
Introduction
Early detection is one of the most important factors to improve treatment and decrease the cancer mortality, especially Lung Cancer (LC), which is highly mortal and aggressive. The LC mortality has dramatically increased in spite of several pharmacological and image technical advances [1,2]. The high incidence of LC has been associated with cigarette smoking, however genetic diversity and environmental pollution must also be considered as risk factors, especially in those cities highly exposed to environmental carcinogens. In Chile, LC has the second high mortality rate/100,000 population of cancer, after stomach cancer. During 2012, in the country the LC incidence for males was 17.1 with a mortality of 17.0, while for females the incidence was a 10.2, with a mortality of 8.8 [3]. In the same period, the Antofagasta region (North of Chile), historically exposed to high levels of drinking water Arsenic (dw-As), showed a mortality rate of 31.6 for both genders, with a rate of 44.2 and 17.4, for men and women, respectively [4-8]. The implementation of earlydetection technologies and prognostic biomarkers are imperative, specifically in cities highly exposed to environmental carcinogens, such as PAHs and Arsenic [9-13]. It is extremely important to have complementary and noninvasive tools, to apply in asymptomatic population, especially to individuals with high risk of LC. In this context, circulatingfree DNA (cfDNA) levels have been described as a potential tool to detect cancer cases (lung, breast, colorectal, other types of tumors) in comparison with controls [14-18]. However, not much is known in relation of cfDNA levels in subjects with Pre-neoplastic Lesions (PNL). Additionally, Copy Number Alterations (CNAs) are amplified or deleted regions of the genome, and have been described as key events in tumor progression and identified in different LC subtypes [19-23]. This study proposed to evaluate cfDNA levels and determine CNAs in serum cfDNA in LC and PNL patients, which have been diagnosed according to various standard tests including Computerized Tomography (CT), Autofluorescence Bronchoscopy (AFB), Automatic Quantitative Cytology (AQC) and a tumor biomarker (Onko-sureTM) protocols, as previously described by Adonis et al. [24].
Methods
Population
The study enrolled a total of 107 people, from the Metropolitan (Santiago city, N=51) and Antofagasta (N=56) regions, with medium or high LC risk, according to a LC Risk Survey, as has been described in Adonis et al. [25]. The enrolling process has been part of the INNOVA CORFO grants (07CN13PBT-48 and 11IDL2-10634, 2012), that included a survey of LC risk and different assays to evaluate the LC risk. The survey included questions related with smoking habit, hereditary cancer history, environmental pollutants exposure and dietary habits. The voluntaries were males and females, aged 40 years or older; with family history of LC, non-smokers and ever smokers exposed naturally to environmental air pollution (Santiago) or to dw-As (Antofagasta) for at least 10 years. According their clinical symptoms, thirty three (33) of the volunteers enrolled were suspected of having LC, with the final diagnosis completed in the study. These patients did not have any treatment at the moment to be included in the study. The voluntaries with medium or high risk of LC were invited to evaluate the LC risk with AQC in induced sputum and with the tumour marker DR70 in serum [24-28]. According to AQC and DR70 results, the voluntaries were classified under three categories of LC risk (low, medium and high). Participants with medium and high LC risk, were invited to have an AFB, and submitted to a CT. Endobronchial mucosal biopsies were taken from all areas that were suspicious under WLB or AFB [29-31]. In addition, surveillance biopsies were taken from epithelium with normal appearance in all subjects. In addition to CT, pathology assays were conducted in the biopsies collected at bronchoscopy. The final diagnosis was confirmed as normal, pre neoplastic lesion (PNL) (metaplasia or dysplasia) or LC. All the voluntaries signed an informed consent.
Serum cfDNA isolation and amplification
Peripheral blood was collected and the serum was carefully separated centrifuging twice at 3,500 rpm for 10 min at room temperature. The serum specimens were stored in 1mL aliquots at -80°C until use for the DR70 assay and cfDNA isolation. The cfDNA was isolated from serum aliquots using Qiagen QIAamp DNA mini kit (Westburg, Leusden, The Netherlands), according to the manufacturer's instructions for blood and body fluids [32-34]. The amount of serum cfDNA was determined in quadruplicate using qPCR, with TaqMan® Copy Number Reference Assay RNase P (Applied BioSystems, Nieuwerkerk a/d IJssel, The Netherlands), according to Life Technologies recommendations. This assay detects the Ribonuclease P RNA component H1 (H1RNA) gene (RPPH1) on chromosome 14, cytoband 14q11.2. The standard calibrator curve was done with wg-DNA from the healthy volunteer obtained with the same system of QIAamp DNA mini kit, describe before. The purified products (cfDNA and wgDNA) were amplified in triplicate with an isothermal method using the Illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences). The triplicate products were pooled to reduce the allele bias and to avoid differential amplification of particular segments of the genome [35,36]. Agarose gel electrophoresis was done for the pooled samples digested with DpnII according to the manufacturer’s instructions (New England BioLabs). The fidelity of the amplification method was confirmed by both allele and CNV specific polymorphism qPCR assays. The AS3MT gene allele was analyzed in duplicate with the TaqMan® SNP Genotyping Assay SNP (ID: rs11191439, Life Technologies) while the CNV assays for DOCK and LMLN (DOCK8 ID: HS02618091; LMLN ID: Hs04763841) were done in quadruplicate according to the manufacturer’s instructions. The RNase P TaqMan® Copy Number Reference Assay was used as control (Life Technologies).
Microarray hybridization
According to the histopathologic analysis, 26 samples were selected, matching cfDNA and gDNA, for the CNV assays by CGH. The samples included 8 healthy voluntaries, 8 PNL patients (6 Met, 2 Dys) and 10 LC patients (4 SqCC, 4 AdC, 2 SCLC). Each pair of samples was labeled with Cy5-dCTP (cfDNA) and Cy3-dCTP (wg-DNA) using the Klenow fragment of DNA polymerase I and co-hybridized onto HEEBO microarrays (Microarrays Inc., USA). After hybridization, microarrays were imaged using a Scan-Array Lite (Perkin Elmer, USA). Fluorescence ratios for array elements were extracted using GENEPIX PRO 6.0 software (Molecular Devices, USA), and were normalized by print-tip Lowes intra and inter arrays with the software DNMAD [37,38].
Determination of CNAs
Through of HMMs analysis, used as a probabilistic model to determine an unknown sequence of states based upon a sequence of observations [37-39], we defined Minimal Common Regions (MCRs) of recurring DNA gains and losses. The HMM algorithm is available at the ADaCGH server of Asterias (https://adacgh.bioinfo.cnio.es/). Significant differences in CNA frequency were evaluated using the Fisher’s exact test. P-values were adjusted. An adjusted P-value 0.05 was used as a significance threshold [40]. Cytoband mapping was obtained from NCBI MapViewer (https://www.ncbi.nlm.nih.gov/mapview/), using the Human genome Build 38 (GRCh38) (annotation release 105). Gene content and genomic locations at MCRs were obtained using the GeneCard Platform (https://www.genecards.org/) [41]. A gene ontology (GO) enrichment analysis of our gene list was done using WebGestalt (https://bioinfo.vanderbilt.edu/webgestalt/) and GOEAST (https://omicslab.genetics.ac.cn/GOEAST/) [42-45], in order to identify significant GO terms. Additionally, genes defined were analyzed with Gene Prospector (https://hugenavigator.net/HuGENavigator/geneProspectorStartPage.do), for a comparative functional analysis, in order to search evidence of genes related with diseases and risk factors associated to cancer or lung cancer.
Statistical analysis
Median serum cfDNA levels between groups (Healthy, PNL and LC), were analyzed with non-parametrical Krustal Wallis testing. Additionally, the diagnostic cut-off value was determined from ROC curve [46]. Sensitivity, specificity, predictive values (both positive and negative) and the Youden’s index were calculated with Epidat 3.1. The performance of cfDNA was analyzed for the study threshold and ROC area under the curve (AUC) was calculated by the trapezoidal rule and the variance estimate as described Zhou [40].
Results
cfDNA Levels: A total of 107 serum samples were used to determine the cfDNA levels from Cancer Free (CF) volunteers (63) and volunteers with diagnosis of PNL (11) (Met:9; Dis:2) and LC (33) (SqCC, n=9; AdC, n=17; SCLC, n=2; Ind, n=5); classified according to AQC, DR70, AFB and histopathology assay.
The volunteers that were confirmed clinically with LC showed significantly higher levels of cfDNA (average, 16.13 ng/mL; range 1.081-113.9 ng/mL) than Cancer Free volunteers (CF) (p<0.01) (average 2.692 ng/mL; range 0.255-8.151 ng/mL) and Pre Neoplastic Lesions (PNL) (p<0.001) (average 1.961 ng/mL; range 0.257-7.225 ng/mL). On the other hands, the cfDNA levels were unable to discriminate with significance between CF and PNL subjects. The cfDNA in CF subjects were similar to subjects with PNL diagnosed previously by AQC, DR70, AFB and histopathology assay (Figure 1).
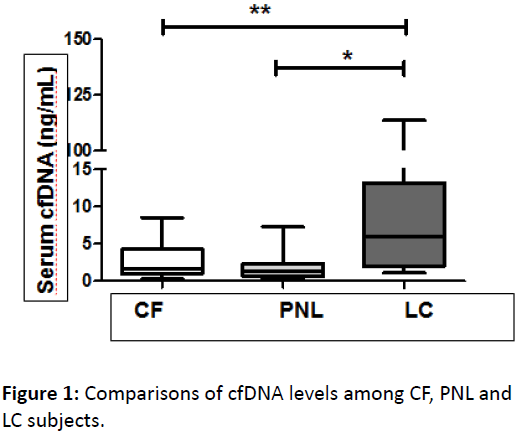
Figure 1: Comparisons of cfDNA levels among CF, PNL and LC subjects.
Box plot displaying cfDNA levels among CF subjects (n=63), subjects with PNL (n=11) and LC (n=33) clinically confirmed. Black lines within the box represent median values, and the whiskers indicate the minimum and maximum values that lie within 1.5x interquartile range. To stratify by region of residence, the LC patients presented significant higher cfDNA levels than the CF subjects either for Metropolitan and Antofagasta region. LC subjects from Antofagasta region showed a non-significant higher cfDNA levels than subjects from the Metropolitan region. In this context, is important to consider that in Antofagasta region a 73% (8) of the LC were SqCC and 18% [2] were AdC. On the other hand, in the Metropolitan region 19% (4) of the cases were SqCC and 67% (4) were AdC. The rest of cases were distributed among SCLC and NSCC-NOS. By stratifying histologically the samples, it was possible to observe that the average cfDNA level for the total sample for SqCC was 17.54 ng/mL while for AdC was 2.67 ng/mL.
Figure 2A shows the overall ROC performance for cfDNA in LC and for PNL (Figure 2B). According to the Youden index, a threshold of 6.075 represent an optimal value, with an empirical specificity of 87.3% (95% IC: 76.5-94.4) and sensitivity of 46.9% (95% IC: 29.1-65.3) for LC with a Predictive Positive Value (PPV) and Predictive Negative Value (PNV), of 65.2% (95% CI: 42.7-83.6%) and 76.4% (95% CI: 64.9-85.6%), respectively. For a more general measure of diagnostic performance for LC, the ROC AUC resulted in a significant value of 0.724 (p<0.0001) (95% CI: 0.623-0.811) (48). For PNL, the test showed a specificity and sensitivity of 72.73% (95% CI: 39-94.0%) and 58.73% (95% CI: 45.6-71.0%); respectively, considering a threshold of 1.5357 according to the Youden index. On the other hand, for PNL the ROC AUC resulted nonsignificant, with a value of 0.604 (95% CI: 0.483-0.716) (p=0.2388).
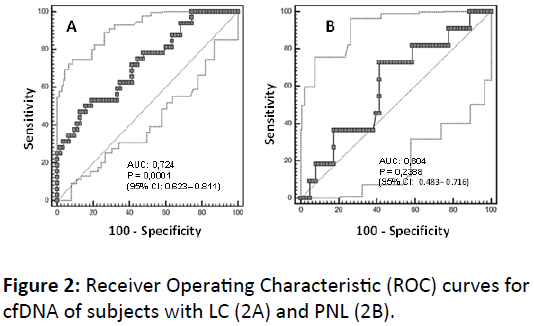
Figure 2: Receiver Operating Characteristic (ROC) curves for cfDNA of subjects with LC (2A) and PNL (2B).
As shown in Figure 3, the CNA pattern of the LC samples was quite different from that of the CF samples. Many CNAs present in the LC samples were not found in the CF samples. However, only four deletions detected in LC sample were significant, respect to CF samples. These loosed regions were located in the chromosomes 2, 7, 11, 17. These alterations were not significantly observed for PNL.
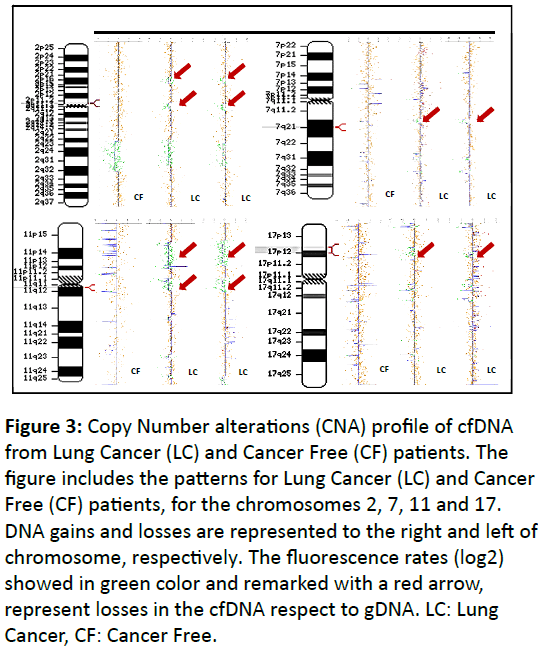
Figure 3: Copy Number alterations (CNA) profile of cfDNA from Lung Cancer (LC) and Cancer Free (CF) patients. The figure includes the patterns for Lung Cancer (LC) and Cancer Free (CF) patients, for the chromosomes 2, 7, 11 and 17. DNA gains and losses are represented to the right and left of chromosome, respectively. The fluorescence rates (log2) showed in green color and remarked with a red arrow, represent losses in the cfDNA respect to gDNA. LC: Lung Cancer, CF: Cancer Free.
The significant losses related with LC were associated with the cytobands 2p11.2-2p11.1, 7q21.2, 11q12.1 and 17p13.1-17p12 and were not associated with smoking habit status or place of residence (Metropolitan or Antofagasta region). The enrichment analysis (WebGestalt and GOEAST) in addition to Gene Prospector, in order to search evidence of genes related with diseases and risk factors associated to cancer or LC, suggested that the genes associated with these alterations are mainly related with immune response, xenobiotic transport, DNA replication and cell proliferation, tumor suppressor, regulation of cell adhesion and spreading and inhibition of cell proliferation.
As is shown in the Table 1, the IGKC and SERPING1 genes were significantly associated to the cytoband 2p11.2-2p11. The IGKC gene codes for the Immunoglobulin Kappa Constant and has been suggested as a prognostic marker for several solid tumor types [50]. The product of the SERPING1 gene has been associated with immune response and tumor development. However, its role in cancer has not been clearly defined. On the other hand, the genes COX10 and ABCB4 were significantly associated with the cytobands in 17p12-17p13 and 7q21.12, respectively.
| Citoband | Genes | Associated Functions | References |
|---|---|---|---|
| 2p11.2 (Ensembl) | IGKC | Immune response | [47,48] |
| 2p12 (Entrez Gene) | |||
| 7q21.12 | DMTF1 | Tumor suppressor | [49] |
| ABCB4 | Xenobiotic transport | [50] | |
| Tumor suppressor in lung cancer | |||
| CTNND1 | Innate immunity | [51] | |
| Epithelial integrity | |||
| 11q12.1 | SERPING1 | Inflammatory response | [52] |
| 17p12 -17p13.1 | COX10 | Response to oxidative stress | [53] |
| MAP2K4 | Tumor suppressor | [54] |
Table 1: Genes whose genomic changes are associated with LC
Additionally, the comparative functional analysis has suggested the DMTF1 gene, located in the fragment 7q21.12 and related with cellular proliferation and cell cycle, as a candidate gene associated to LC. The analysis also suggested to the genes MAP2K4 located in the segments 17p13.1-17p12 and CTNND1 for 11q12.1, both related with cell adhesion. These results suggest that most of the fragments with ANC contain genes associated to a variety of cellular processes such as proliferation, differentiation, adhesion and immune response, and xenobiotic detoxification (Table 1).
Discussion and Conclusion
The aims of this study were to evaluate the levels of cfDNA in serum and their significant CNAs, in order to explore their potential as biomarkers of malignance, in Lung Cancer (LC) subjects, in individuals with Pre Neoplastic Lesions (PNL) and in healthy people exposed to environmental pollutants and high risk of LC.
According our findings, the cfDNA levels in serum increased during malignancy of LC but not significantly during premalignant lesions or pre-invasive stages as reported previously [33]. None of the subjects with pre-invasive lesions, either metaplasia or dysplasia was discriminated from controls. LC subjects showed significantly higher cfDNA levels than cancer free subjects, as was previously described [33].
The ROC curve showed that the cfDNA provides significant discrimination for LC over a wide range of score thresholds. The threshold evaluated according to the Youden index (6.075), showed a sensitivity and specificity of 46.9% and 87.3%, respectively. The PPV and PNV, were 65.2% (95% CI: 42.7-83.6%) and 76.4% (95% CI: 64.9-85.6%), respectively. According to the results, cfDNA might would support in the detection and confirmation of positive LC cases and especially discard the presence of malignancy, with high precision (specificity of 87.3% and PNV of 76.4%). According to the area under the curve (AUC) of the cfDNA was 0.724 (95% CI: 0.623-0.811), suggesting a fair ability of cfDNA for LC diagnosis.
Additionally, detection of cancer-related molecular alterations in cfDNA may provide a complementary advantageous tool for diagnosis of LC. We show evidence that the amplification procedure of cfDNA did not alter significantly the DNA copy number composition of cfDNA samples. Therefore, our results suggest that the cfDNA from LC patients contain specific genome alterations respect to the controls or the healthy volunteers. Our analysis showed that candidate genes associated to LC are located within the regions with significant CNV, mainly deletions. The genes deleted and associated with LC or with the tumorigenesis process, are related mainly with immune response, inflammation and cellular processes as well as proliferation, differentiation and apoptosis.
Two of the candidate genes, IGKC and SERPING1, are associated with immune response. The IGKC gene, has been associated with a longer survival for LC, mainly for AdC [50], suggesting that the humoral immune response plays an important role in non-small cell lung cancer [47,48]. It has been described that IGKC expression, is an adaptive humoral immune response and its effect is host-dependent and might be related with the capacity of the tumor cells to produce immunoglobulin against tumor cells [48,49]. Most of the studies of immune response against tumor cells have been focused to cytotoxic response mediated by T cells, but not by the B cells. It has been described that a suppressed cellular immunity in a chronic inflammatory process could promote risk to development several cancers [49,50]. The immune system is an extremely complex system that is still not fully understood, however, these findings might suggest a role of the humoral anti-tumor response, showing that anti-bodies, as IGKC, against tumor might takes place within the tumor. A deletion of the IGKC gene could be related with a lower survival of patients host and promote growth and survival of tumor cells.
On the other hand, when the tissues are exposed to chemical irritant or xenobiotic, inflammatory cells are implicated in removing damaged cells promoting cell death and phagocytic pathways as well as any metabolic process and cell proliferation that allow tissue regeneration and damage repair. Obviously, the inflammation process shall subside after damage removing or tissue regeneration and not produce a chronic inflammation process. Today, it is well accepted that an inflammatory microenvironment is an essential component of all tumors and many environments or cancer risk factors could be associated with to promote some form of chronic inflammation and contribute with the cancer development [51,52].
On the other hand, CTNND1 (p120 catenin), has been described as an important component of lung epithelial cells (flat type I cells). The lungs are permanently exposed to exogenous compounds requiring protection and defense mechanisms. The p120 gene, has shown an important role regulating cadherin stability and innate immunity [53-55] and its deletion or depletion has been related with loss of the alveolar epithelial integrity. A loss of epithelial integrity could be related with absence of p120 that prevents the internalization and degradation of E-cadherin. Then p120 deletion could induce a defective epithelial barrier that in genotoxic or pathogens exposition might promotes a chronic inflammation process. Additionally, p120 has an important role in surfactant proteins production (Sp-B, Sp-C, and Sp-D), that play a protective role against lung infection, allergy, asthma, and inflammation by modulating host defense and inflammation [56].
In this point of view, a deletion of SERPING1 (C1 inhibitor, C1-INH) gene, that encodes a protein involved in the regulation of the complement cascade (C1 inhibitor), could be related with a major cancer risk. The complement needs a strict control and the absence of SERPING1 conduces to a spontaneous and permanent activation of C1, stimulating vessel growth and a chronic inflammation or an altered inflammatory response [57].
A deletion of SERPING1 leads to a loss of inhibition of C1s action, inducing excessive levels of C3a and C5 and thereby an increase of the vascular permeability. Additionally, the absence of SERPING1 contributes to a permanent activation of Kallikrein (KLK1) and XIIa coagulation factor (Hageman factor), that promotes an excessive generation of bradykinin a potent vasodilator peptide, activating the bradykinin B2 receptor. KLK1 is consistently elevated in the bronchial alveolar lavage fluid of individuals with asthma or chronic bronchitis, particularly after stimulation by allergens or Broncho obstructive agents [58,59].
A permanent activation of KLK1 means also an activation of pro-epidermal growth factor (pro-EGF) in human airway goblet cells to release mature EGF, which activates EGF receptor (EGFR), resulting in mucus hypersecretion and goblet-cell hyperplasia [59].
Additionally, our study suggests DMTF1 (Dmp1) as a candidate gene associated to cfDNA and as tumor suppressor. The DMTF1 is homozygous deleted in approximately 40% of human non-small-cell lung cancer and has been proposed as a potential prognostic and gene-therapy target for non-smallcell lung cancer [60]. The Dmp1 promoter is activated by the oncogenic Ras-Raf-MEK-ERK-Jun pathway but is repressed by overexpression of c-Myc, E2Fs, and by physiological mitogenic signaling. Dmp1 prevents tumor formation by activating the Arf-p53 pathway [61] and promoting apoptosis and cell cycle arrest in primary cells. In the same context of tumor suppressor, the MAP2K4 loss, has been described as a tumor suppressor in lung adenocarcinomas and inhibits tumor cell invasion by decreasing peroxisomal proliferator-activated receptor γ (PPARγ2) levels, which has been associated with different tumor models [62]. In conclusion, the Masp2k4 deletion could be related with an increased growth of lung neoplasia, and promotion of invasion and metastasis in both early and advanced stages of lung tumorigenesis.
In agreement with the suppressor role of the 7q21.12 segment in lung cancer, this study suggests that the ABCB4 gene (MDR3) could be related to LC. Although the exact function of ABCB 4 in lung epithelial cells and LC has not been well defined, would be possible since its function as a transporter of a wide variety of substrates across biological membranes, to hypothesize a crucial role in the protection against xenobiotics. Additionally, it has been described that an over-expression of ABCB4 significantly suppresses the formation and proliferation of lung cancer cells. A frequently silenced ABCB1 in different cancers has been observed suggesting that this gene may act as a tumor suppressor in lung cancer [63].
Finally, COX10 (Cytochrome c oxidase), is the terminal component of the mitochondrial respiratory chain and catalyzes the electron transfer from reduced cytochrome c to oxygen. It has been described that hypoxia occurs in most solid tumors, and malignant progression and poor prognosis have been related with a hypoxia state [64,65]. It has been described that inside the primary tumor, the rapid proliferation of cancer cells, frequently leave many tumor cells in regions where the oxygen concentration is significantly lower compared with normal tissues. To survive in this hypoxic microenvironment, cancer cells need to change their intrinsic gene expression, probably by inducing angiogenesis to provide enough nutrients and oxygen [66].
As mentioned, tumor cell adapts their mitochondrial metabolism regulation by increasing glycolysis and suppressing the expression of iron-sulfur cluster scaffold homolog (ISCU) and cytochrome c oxidase assembly protein (COX10) [67,68]. The COX deficiency or deletion has been described in mitochondrial diseases, as response to oxidative stress [67].
In conclusion, cfDNA levels in serum increased during malignancy of LC but not significantly during pre-malignant lesions or pre-invasive stages. Our results support the idea that cfDNA could be used as a complementary tools to prognosis and evolution of LC, with high precision (specificity of 87.3% and PNV of 76.4%) [69].
The small DNA fragments, called cfDNA, are released into the circulation as a consequence of necrosis, apoptosis and/or active secretion from cells in a tumorigenesis or inflammation process [70-72]. These genomic fragments could be present in cancer free patients, but in cancer patients a significant fraction shall come from tumor cells [72]. Then, chromosomal aberrations in cfDNA may reflect CNAs arising during the tumorigenesis process, as has been described previously [73,74].
This study suggests that cfDNA carry some chromosomal aberrations related with the lung tumorigenesis process that could be used to confirm a suspected case of LC, prior to indicate more complex or expensive exams. Within these alterations our results suggest the loss of specific chromosomic fragments, contain genes related with inflammation response (SERPING1), humoral immune response (IGCK), innate immunity and epithelial integrity (CTNN1), tumor suppressor (MAP2K4, DMTF1 and ABCB4), and response to oxidative stress (COX10). Although, these genes will need to be validated and confirmed, in order to evaluate whether candidate genes and copy number alterations have a real prognostic value, our results suggest that the cfDNA and its chromosomal aberrations could have a potential as LC diagnostic biomarkers.
These genomic markers could be used as complementary tools in order to improve the diagnosis of LC and mainly diminish the invasive or expensive procedures, allowing selecting the patients candidates to additional exams [18,60,69]. Additionally, these biomarkers could have a great potential as screening methods in population with high risk for LC.
Acknowledgments
This work was supported by Grants from INNOVA-CORFO Chile (07CN13PBT-48 and 11IDL2-10634, 2012). We also thank all the volunteers that accepted to participate in these projects. This work has been carried out with ethical committee approval of the Faculty of Medicine of University of Chile.