Keywords
COVID-19, SARS-CoV-2 variant/mutant, Spike protein (S), D614G, Tracking
alga- rhythms, Predictive stewardship
Introduction
Coronavirus family has caused several human illnesses, the
latest caused by SARS-CoV-2, has led to COVID-19 pandemic
posing serious threat to global health [1-3]. A SARS-CoV-2 variant
encoding a D614G mutation in the viral spike (S) protein has
now become the most prevalent form of the virus worldwide,
suggesting fitness advantage for the mutant [4-7]. The G614
variant is associated with higher upper respiratory tract viral load,
higher infectivity, increased total S protein incorporation into the
virion, reduced S1 shedding and a conformational change leading
to a more ACE2- binding and fusion- competent state. However,
it does not seem to be correlated to increased disease severity
or escape neutralizing antibodies [8-12]. The SARS-CoV-2 D614G
S protein variant sup- planted the ancestral virus worldwide,
reaching near fixation in a matter of months. Here it is shown that
D614G was more infectious than the ancestral form on human
lung cells, colon cells, and on cells rendered permissive by ectopic
expression of human ACE2 or of ACE2 orthologs from various
mammals. D614G did not alter S protein synthesis, processing,
or incorporation into SARS-CoV-2 particles, but D614G affinity
for ACE2 was reduced due to a faster dissociation rate [13,14].
Consistent with this more open conformation, neutralization
potency of antibodies targeting the S pro- tein receptor-binding
domain was not attenuated. Over the course of the SARS-CoV-2
pandemic, the identified SNPs were seen only once in the dataset, and only four SNPs rose to high frequency reported in
GISAID [15,16]. This suggests that D614G confers replication
advantage to SARS- CoV-2 increases the likelihood of humanto-
human transmission, which would support this hypothesis.
Future prospective comparisons of D614G transmission to that
of D614 seem unlikely given that D614G has gone to near fixation
worldwide. However, the SARS-CoV-2 genomes that have been
sequenced are only a narrow snapshot of the pandemic and
additional sequencing of archived samples might pinpoint the
origin of D614G or better resolve the variant’s trajectory. This
D614G is associated with increased viral load in people with
COVID-19 although these studies quantitated SARS-CoV-2 RNA
and did not measure infectious virus.
When the SARS-CoV-2 S protein RBD is in its closed conformation,
the binding site for ACE2 is physically blocked [17]. Models of
coronavirus S-mediated membrane fusion describe ACE2 binding
to all three RBD domains in the open conformation as destabilizing
the pre-fusion S trimer, leading to dissociation of S1 from S2 and
promoting transition to the post-fusion conformation.
According to these models, the well-populated all- open
conformation of D614G would reflect an intermediate that is on-pathway to S-mediated membrane fusion [18]. Therefore,
structural biology provides key insights into 3D structures,
critical residues/mutations in SARS-CoV-2 proteins, implicated
in infectivity, molecular recognition and susceptibility to a broad
range of host species [19].
The detailed understanding of viral proteins and their complexes
with host receptors and candidate epitope/lead compounds is the
key to developing a structure-guided therapeutic design. Since
the discovery of SARS-CoV-2, several structures of its proteins
have been determined experimentally at an unprecedented
speed and deposited in the Protein Data Bank [20,21]. Further,
specialized structural bioinformatics tools and resources have
been developed for theoretical models, data on protein dynamics
from computer simulations, impact of variants/mutations and
molecular therapeutics. Here, we provide an overview of ongoing
efforts on developing structural bioinformatics algorithms and
predictions for COVID-19 research. We also discuss the impact
of these resources and structure-based studies, to understand
various aspects of SARS-CoV-2 variant infection and therapeutic
development. These include understanding differences between
SARS-CoV-2 and SARS- CoV-2 variant, leading to increased
infectivity of SARS-CoV-2, and deciphering key residues in the
SARS-CoV-2 involved in variant as well.
Methods
Coronavirus sequences and structures analysis
Amino acid sequences and mutant data of the S protein used in
the analysis were obtained from NCBI GenBank and GISAID. After
the first complete genome sequence of SARS-CoV-2 released on
NCBI GenBank (accession number: NC 045512.2), there has been
a large number of genome sequences as proceeding information
is referred in Table 1. The mutant information of whole- genome
sequences of S protein with high coverage of SARS-CoV-2 strains
from the infected individuals around the world was obtained from
the GISAID database (https://www.gisaid.org). Sequence analysis
and k-means clustering were described in detail elsewhere.
For structural analyses, visualization, analysis and in silico
mutations of protein structures were done (see Table 2). First, we
downloaded the molecular structure of the spike protein from
the Protein Data Bank (PDB). This structure corresponds to that
resolved by colleagues and deposited in PDB .
Table 1: The SARS-CoV-2 proteome (NCBI reference genome NC_045512.2)
| Gene |
Protein
length |
Position in Genome |
Description |
| Nsp1 |
180 |
266-805 |
Interferes with host mRNA translation and processing. |
| Nsp2 |
638 |
806-2719 |
Specific function is not known, it may play an auxiliary role to other viral proteins. |
| Nsp3 |
1945 |
2720-8554 |
Papain-like protease with phosphatase activity. Performs proteolytic cleavage of the polyproteins, membrane arrangements |
| Nsp4 |
500 |
8555-10054 |
Involved in membrane rearrangements during viral infection. |
| Nsp5 |
306 |
10055-10972 |
3C-like proteinase that cleave the viral polyprotein to produce the active forms of the nonstructural proteins. |
| Nsp6 |
290 |
10973-11842 |
Involved in membrane rearrangements during viral infection and autophagy. |
| Nsp7 |
83 |
11843-12091 |
Forms an hexadecameric complex with nsp8 that helps in viral RNA replication. |
| Nsp8 |
198 |
12092-12685 |
Forms an hexadecameric complex with nsp8 that helps in viral RNA replication. |
| Nsp9 |
113 |
12686-13024 |
Binds and protects the viral genome from host degradation during replication. |
| Nsp10 |
139 |
13025-13441 |
Interacts with nsp14 and nsp16 to perform 3′–5′ exoribonuclease and 2′-O-methyltransferase activities, respectively. |
| Nsp11 |
13 |
13442-13480 |
Short peptide with potential role in RNA synthesis. |
| Nsp12 |
932 |
13442-16236 |
RNA-dependent RNA polymerase. |
| Nsp13 |
601 |
16237-18039 |
Viral RNA helicase. |
| Nsp14 |
527 |
18040-19620 |
3′-to-5′ exonuclease with proofreading activity. |
| Nsp15 |
346 |
19621-20658 |
Nidoviral RNA uridylate-specific endoribonuclease (NendoU) |
| Nsp16 |
298 |
20659-21552 |
2′-O-ribose methyltransferase. Involved in capping of viral mRNA to protect it from host degradation. |
| S |
1273 |
21563-25384 |
Spike glycoprotein. Interacts with human ACE2 to enter target cells |
| M |
222 |
26523-27191 |
Membrane glycoprotein. Required for viral particle assembly. |
| N |
419 |
28274-29533 |
Nucleocapsid protein. Binds viral RNA during viral particle formation. |
| E |
75 |
26245-26472 |
gEnvelope protein. Forms ion channels in host ER membranes.
Involved in exaggerated immune response. |
| ORF3a |
275 |
25393-26220 |
Form ion channels in the host membrane. Linked to inflammatory, IFN signaling, innate immunity, apoptosis, and cell cycle regulation. |
| ORF6 |
61 |
27202-27387 |
Viral replication enhancer. |
| ORF7a |
121 |
27394-27759 |
Viral replication enhancer. Prevents virus tethering at the plasma membrane by inactivation BTS-2 protein. |
| ORF7b |
43 |
27756-27887 |
gEnvelope protein. Forms ion channels in host ER membranes.
Involved in exaggerated immune response. |
| ORF8 |
121 |
27894-28259 |
Virus replication enhancer. |
| ORF9b* |
97 |
28284-28580 |
Expressed from an alternative reading frame in the N gene. Suppresses host antiviral responses by promoting MAVS degradation . |
| ORF10 |
38 |
29558-29674 |
Potential role in hijacking components of the host ubiquitin- proteasome system (UPS) |
| ORF14** |
73 |
28734-28946 |
Expressed from an alternative reading frame in the N gene. Unknown function. |
Table 2: Oligonucleotide primers used for amplification of SARS-CoV-2 nucleoprotein gene, a single point mutation in the N gene.
| Type |
Name |
Sequence |
Remark |
| N2-Probe |
Probe |
ACAATTTGCCCCCAGCGCTTCAG |
|
| N2-FP |
Control |
TTACAAACATTGGCCGCAAA |
|
| |
27870fwd |
GAAACTTGTCACGCCTAAACGAAC |
|
| |
28268fwd |
ACTAAAATGTCTGATAATGGACC |
|
| |
28923fwd |
CTGCTCTTGCTTTGCTGCTGC |
|
| |
29338fwd |
GCATATTGACGCATACAAAAC |
|
| N2-RP |
Control |
TTCTTCGGAATGTCGCGCA |
|
| |
28943rev |
GCAGCAGCAAAGCAAGAGCAG |
|
| |
29358rev |
GTTTTGTATGCGTCAATATGC |
|
| |
29588rev |
AGCGAAAACGTTTATATAGCCCATCTG |
|
| |
29880pArev |
TTTTTTTTTTGTCATTCTCCTAAGAAGCTAT T |
|
| SNP |
C28858T |
|
N-Nterm |
| |
C29200T |
|
N-mid |
| |
C29451T |
|
N-Cterm |
*Note: Primer names reflect the 5’ end of the respective oligonucleotide on the reference genome NC_045512.2.
Genetic algorithm (GA) application
A population with defined fitness undergoes random mutation,
crossover, and selection, and those with high fitness are retained.
The individuals in the population are RNA sequences in our case
and the fitness function is the number of residues that have
the same 2D structure in both the target (provided for fitness
calculation) and the predicted structure as determined by the
Hamming distance, a metric for comparing two binary data
strings. Based on prediction performance on the residue FSE
(Frame shifting RNA Element) and computational complexity (see
below result section), we choose NUPACK [22] for our GA [23,24].
The Nucleic Acid Package web server (http:// www.nupack.org) currently enables and GAs mimic evolution in nature. The initial
population is obtained by randomly assigning nucleotides to the
mutation region in the RNA sequence. This population is then
subject to iterations of random mutation, crossover, selection,
and nominatio.
Nucleotide and amino acid variant detection
We first aligned each of these SARS-CoV-2 sequences using BLAT
software [25,26]. After the alignment, we extracted nucleotide
sequences corresponding to individual proteins of SARS-CoV-2,
translated them to amino acid sequences, and then compared
them to reference amino acid sequences. Using the nucleotide
mutations, the resulting amino acid mutations throughout the
proteome of SARS‐CoV‐2 were determined. The amino acid
changes were automatically annotated using the bioinformatics
tool and a subsequent run, the resulting proteome from each
SARS‐CoV‐2 genome was created and edited using CLC Genomics
Workbench 20.0.3 [27]. This bioinformatics software is used by
hundreds of microbiology and virology labs around the world for
basic research and infectious disease epidemiology. The whole
proteome was then aligned for phylogenetic analysis, and for
identification of the resulting amino acid mutations.
Energy calculation
The effects of mutations on protein stability of S and binding
affinity of RBD with hACE2 were estimated [28] by the folding
energy change (G) and the binding energy change (G) between
the mutant structure (MUT) and wild-type (WT) structure,
respectively [29]. FoldX was used for energy calculations [30,31].
The performance of the FoldX compares favourably with other
random- based approaches for protein engineering research
including therapeutic antibody design. Particularly, FoldX is widely
used for computational saturation mutagenesis in biomedical
studies. All protein structures were repaired and stability analysis
was performed. The folding energy change was calculated using
an Equation
ΔΔG stability = ΔG folding MUT − ΔG folding WT
A negative ΔΔG value suggests that the mutation can stabilize
the protein and a positive value indi- cates that it makes the
protein unstable. The structure-based tools DUET and CUPSAT
were applied to check the reliability of FoldX for protein stability
predictions [32,33]. Additionally, interaction analysis carried out and
the binding energy change was computed by an Equation [34].
ΔΔΔG binding = ΔΔG binding MUT − ΔΔG binding WT
A negative ΔΔΔG value suggests that the mutation strengthens
the binding affinity, whereas a positive value indicates that the
mutation weakens the RBD–ACE2 interaction.
TopNetTree model for PPI BFE changes upon
mutation
TopNetTree is a recently developed deep learning algorithm
that integrates the advantages of convolutional neural networks
and gradient-boosting trees [35]. The topology-based network tree (TopNetTree) was constructed by an innovative integration
between the topological representation and NetTree for
predicting PPI BFE changes following mutation ΔΔG [36,37]. In
this work, Top NetTree is applied to predict the BFE changes of
mutations that happened on the RBD with ACE2 of SARS-CoV-2.
The topology-based feature generation is the first step followed
by a convolutional neural network- assisted model as described in
more details below result section. The topological representation
uses element- and site-specific persistent homology to simplify
the structural complexity of protein–protein complexes and
encode vital biological information into topological in- variants.
LSTM Network Based on NLP and Infection Rate
We propose a deep learning model based on Long Short-Term
Memory (LSTM) for sentiment classification of COVID-19–related
comments, which produces better results compared with several
other well-known machine-learning methods [38,39]. Deep
neural networks have the capacity to fit complex distributions
but tend to overfit without sufficient supervision. As infection
rate features are based on the growing percentage of each factor,
they are stable across time. However, epidemic models based on
the infection rate cannot predict policy changes and emergency
conditions nor ad- just the prediction with short-term influence.
Therefore, we introduce the LSTM network based on Neurolinguistic
programming (NLP) features to model the current policy
and social media. Then, the short-term flexibility and long-term
stability are both ensured.
Statistical data analysis
Fisher’s exact test was used to analyze the enrichment of
epitopes and differences of mutation rates of SARS-CoV-2
isolated from different areas. Statistical analysis was carried
out using the R statistical environment version 3.6.1. Mutation
hotspots were identified as genome sites with two or more
occurring mutations; on the other hand, mutation cold spots
are those with no occurring mutations. The characterization of
nucleotide mutations was done in terms of the nature of the
nucleotide substitution (transition or transversion) and insertion
and deletions (indel). The mutation densities ( see an Equation) in
the genome and proteome of SARS‐CoV‐2 were determined [40].
Mutation density = number of mutations ÷ size of genomic (nt
length) or proteomic (aa length) region
Bioinformatic analysis: Data quality control and
processing
Read quality control was carried out using FAST-QC and the
default parameters [41]. Adapter sequences and low-quality
bases were removed. Low-complexity reads, those with a length
shorter than 40 bases, and duplicates were excluded using CDHIT-
DUP v.4.6.8, where CD-HIT is a widely used program for
clustering biological sequences to reduce sequence redundancy
and improve the performance of other sequence analyses. Offtarget
reads were then filtered out using Bowtie2 v2.3.4.3 with
the default parameters against human genome version GRCh38.
p13 (Genome Reference Consortium Human Build 38 patch release 13), and the SILVA (a ribosomal RNA) database as a
reference to filter out human DNA and ribosomal sequences [42].
Results
It is reported that increased COVID-19 pandemic severity has not
been detected in association with D614G infection [15, 43-45].
Perhaps there are fitness tradeoffs for D614G in vivo due to the
more open conformation of its RBD, which potentially renders
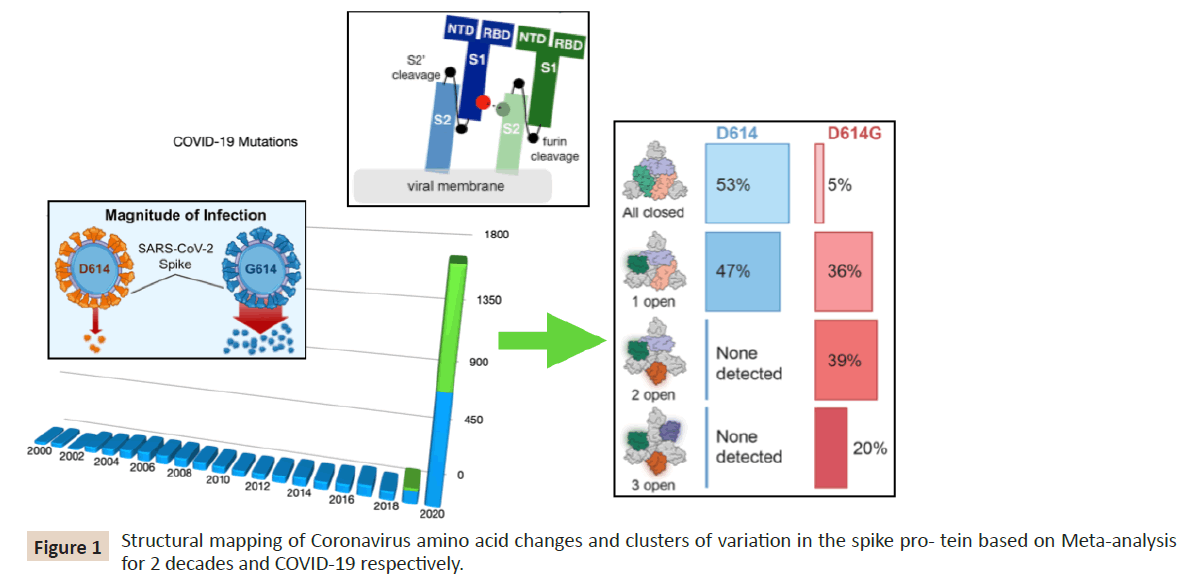
D614G more immunogenic [44, 46-48]. As shown in (Figure 1)
coronavirus mutants are prevalent since the end of 2019 and
the location of D614G within the S protein is remote from the
receptor-binding domain in keeping with the fact, that D614G
affinity for ACE2 is less than that of D614, and that the relatively
better-concealed D614 receptor-binding domain is likely to be
advantageous for immune evasion, the D614G and D614 variants
are equally sensitive to neutralization by human monoclonal
antibodies targeting the S protein RBD. If SARS-CoV-2 Spike D614G
is an adaptive variant that was selected for increased human-tohuman
transmission after spillover from an animal reservoir, one
might expect that in- creased infectivity would only be evident
on cells bearing ACE2 orthologs similar to that in humans. The
increased infectivity of D614G was equally evident on cells bearing
ACE2 orthologs from a range of mammalian species. Among these
viruses, only SARS-CoV-2 possesses a polybasic furin cleavage site
at the S1-S2 junction in the S protein, which is required for SARSCoV-
2 to in- fect human lung cells but not other cell types (Figure
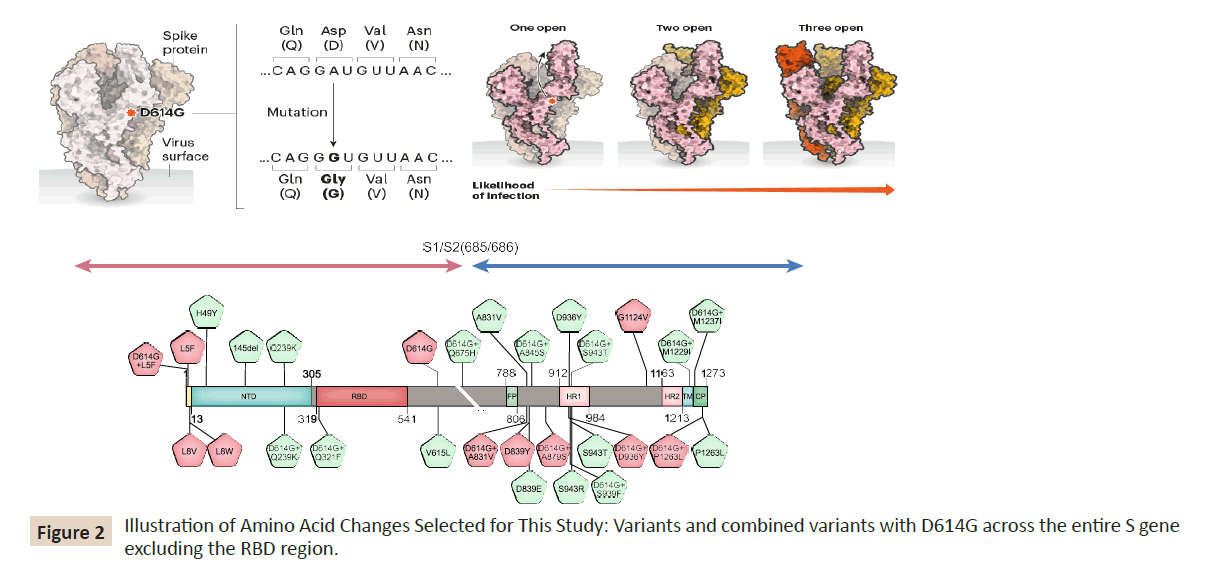
1). The mutation that loosens the spike protein is illustrated in
(Figure 2) Spike proteins on SARA-CoV-2 bind to receptors on
human cells helping the virus to enter. A spike protein is made up
of three smaller peptides in open or closed orientations; when
more are open. It’s easier for the protein to bind. The D614G
mutation, the results of a single-letter change to the viral RNA
code, seems to relax connections between peptides. This makes
open conformations more likely and might increase the chance of
infection as shown in the top of (Figure 2). Further, variants and
combined variants with D614G across the entire S gene excluding
the RBD region are more detailed in the bottom of (Figure 2).
Figure 1: Structural mapping of Coronavirus amino acid changes and clusters of variation in the spike pro- tein based on Meta-analysis
for 2 decades and COVID-19 respectively.
Figure 2: Illustration of Amino Acid Changes Selected for This Study: Variants and combined variants with D614G across the entire S gene
excluding the RBD region.
Presence of a novel mutation and a high
frequency mutation in SARS‐CoV‐2
SARS-CoV-2 is a RNA coronavirus responsible for the COVID-19
pandemic of the Severe Acute Respiratory Syndrome. RNA
viruses are characterized by a high mutation rate, up to a million
times higher than that of their hosts. The virus mutagenic
capability depends upon several factors, including the fidelity
of viral enzymes that replicate nucleic acids, as SARS-CoV-2 RNA
dependent RNA polymerase (RdRp). Mutation rate drives viral
evolution and genome variability, thereby enabling viruses to
escape host immunity and to develop drug resistance (Figure 3).
Figure 3: Neutralization of SARS-CoV-2 RBD mutants by monoclonal antibodies and position of substitutions conferring neutralization.
The morbidity of SARS-CoV-2 (COVID-19) is a serious public
health concern globally and it is enigmatic how several antiviral
and antibody treatments were not effective in the different
period across the globe [49]. With the drastic increasing number
of positive cases around the world WHO raised the importance in the assessment of the risk of spread and understanding genetic
modifications that could have occurred in the SARS-CoV-2. Using
all available deep sequencing data of complete genome from all
over the world (NCBI repository).
In the present work we have compared the SARS-CoV-2 reference
genome to those exported from the GISAID database with the
aim of gaining important insights into virus mutations, them
occurrence over time and within different geographic areas. A
total of SARS-CoV-2 sequences were collected from NCBI and
GISAID databases and aligned against SARS-CoV-2 reference
sequence NC_045512.2 as presented in Tables 1 and 3.
Table 3: Conserved mutations in SARS-CoV-2 genome.
| Mutations |
Amino Acid Change |
Gene |
Remark on Type |
| C to U—n241 |
- |
5′ UTR |
Non coding |
| C to U—nt313 |
No (L16) |
Nsp1 |
Synonymous |
| C to U—nt1059 |
T85I |
Nsp2 |
Missense |
| G to A—nt1397 |
V198I |
Missense |
| Deletion 1606–1609 |
D268 deletion |
Missense |
| C to U—nt3037 |
No (F106) |
Nsp3 |
Synonymous |
| C to U—nt8782 |
No (S76) |
Nsp4 |
Synonymous |
| C to U—9802 |
No (A416) |
Synonymous |
| G to U—9803 |
No (A417) |
Synonymous |
| G to U—nt11083 |
L37F |
Nsp6 |
Missense |
| C to U—nt14408 |
P232L |
Nsp12 |
Missense |
| C to U—nt14805 |
No (Y455) |
Synonymous |
| U to C—nt17247 |
No (R337) |
Nsp13 |
Synonymous |
| A to G—nt23403 |
D614G |
S |
D614 Missense |
| C to U—nt24034 |
No (N824) |
Synonymous |
| G to U—nt25563 |
Q57H |
ORF3a |
Missense |
| G to U—nt26144 |
G251V |
mRNA targeting |
| C to U—nt27964 |
S24L |
ORF8 |
Missense |
| U to C- nt28144 |
L84S |
Missense |
| C to U—nt28311 |
P13L |
N |
Missense |
| U to C—nt28688 |
No (L139) |
Synonymous |
| GGG to AAC—nt28881-28884 |
R203K and G204R |
Missense |
| G to U—nt29742 |
- |
3′ UTR |
Non coding |
Additionally, Table 2 listed the used primmer for sequencing with
the related descriptions.
Structural and function impact of mutations
Upon viral infection, the viral proteins express in the infected cells
and are processed into small peptides by proteosomes [50]. These
peptides are then presented by HLA molecules on the surface of
the infected cells and recognized by T cells through their T cell
receptors. Thus, the potential T cell epitopes can be derived from
any of the viral structural and non structural proteins. Nucleotide
conservation Shannon entropy is a measure of the amount of information (measure of uncertainty). Conservation of each
of the four nucleotides has been determined using Shannon
entropy. Note that it is assumed log (0) = 0 for smooth calculation
of the SE. For a given sequence of length l, the conservation SE
(converse) is calculated as follows using an Equation:
Conv_SE = -um i=1-4 pNilog4 (pNi)
where pNi = fi /i; fi represents the of represents the occurrence frequency of a nucleotide Ni in the given sequence [51].
Functional importance of the D614G mutation in
the SARS-CoV-2 spike protein
Point mutations in SARS‐CoV‐2 variants: We performed
phylogenetic network analysis using the sequences published in
GISAID to investigate the frequency of point mutations in SARSCoV-
2 variants [52-55]. These sequences were collected and
point mutation were calculated by the phylogenetic network
analysis. We also analyzed the locations of these point mutations
and observed a higher frequency of point mutations in several
locations. In addition to it, we further counted the number of point
mutations per gene in order to further analyze the polarization of point mutations in each gene and found more point mutations in
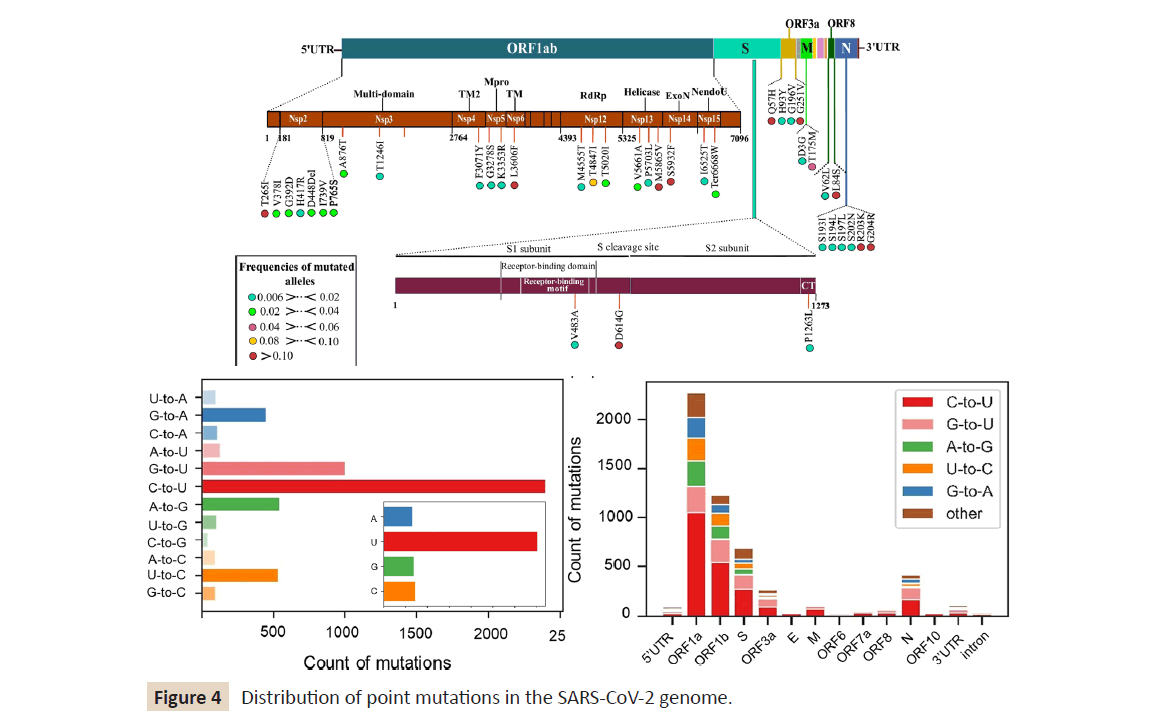
ORF-1a and ORF-1b. However, as shown in (Figure 4) open reading
frame (ORF)-1a and ORF-1b are much longer than other regions,
which may result in more mutations; hence, we estimated the
rate of point mutations per 100 bases in each gene (Figure 4).
When normalized by gene length, the highest frequency of point
mutations occurred in the 5′-untranslated region (UTR) and 3′-
UTR. These results indicate that point mutations are present in
SARS- CoV-2 variants but they do not cluster within the gene
coding regions.
Figure 4: Distribution of point mutations in the SARS-CoV-2 genome.
The Impact on viral infectivity and antigenicity: Severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2) is an enveloped
virus which binds its cellular receptor angiotensin-converting enzyme 2 (ACE2) and enters hosts cells through the action of its
spike (S) glycoprotein displayed on the surface of the SARS-CoV-2.
Compared to the reference strain of SARS- CoV-2, the majority
of currently circulating isolates possess an S protein variant
characterized by an aspartic acid-to-glycine substitution at amino
acid position 614 (D614G). Residue 614 lies outside the receptor
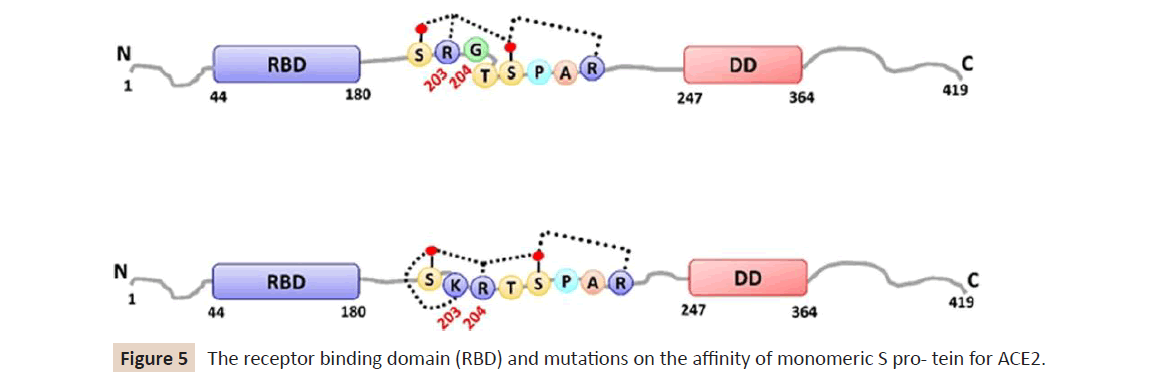
binding domain (RBD) (Figure 5) and the mutation does not alter
the affinity of monomeric S protein for ACE2. However, S(G614),
compared to S(D614), mediates more efficient ACE2-mediated
transduction of cells by S-pseudo-typed vectors and more
efficient infection of cells by live SARS-CoV-2. This review article
summarizes and synthesizes the epidemiological and functional
observations of the D614G spike mutation, with focus on the
biochemical and cell-biological impact of this mutation and its
consequences for S protein function. We further discuss the
significance of these recent findings in the context of the current
global pandemic. The spike protein of SARS-CoV-2 has been
undergoing mutations and is highly glycosylated. It is critically
important to investigate the biological significance of these
mutations. Here, we investigated 80 variants and 26 glycosylation
site modifications for the infectivity and re- activity to a panel
of neutralizing antibodies from convalescent patients [56-59].
D614G, along with several variants containing both D614G and
another amino acid change were significantly more infectious.
Most variants with amino acid change at receptor binding
domain were less infectious, but some variants became resistant
to some neutralizing antibodies. These findings could be of value
in the development of incrementally modified vaccine and future
therapeutic antibodies to quench COVID-19 pandemic eventually.
Figure 5: The receptor binding domain (RBD) and mutations on the affinity of monomeric S pro- tein for ACE2.
Predicting the affinity of ACE2 mutants to SARSCoV-
2 S1 protein using fast methods
We predicted the effect of the detected mutations in ACE2 on the
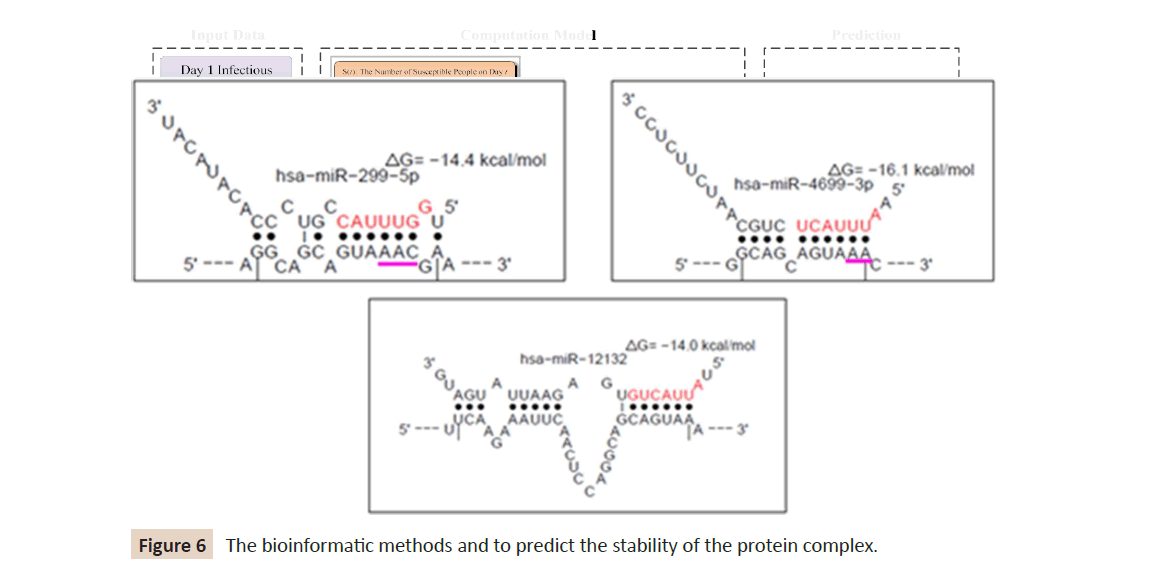
affinity of ACE2 variants to S1 by using different computational methods in this section. There are many bioinformatics methods
to predict the stability of the protein complex and the affinity
between subunits by using various approaches as shown in
(Figure 6). The possible source of the variation between the
prediction results of thermodynamic-based and other descriptorbased
affinity predictors is the interface issue.
Figure 6: The bioinformatic methods and to predict the stability of the protein complex.
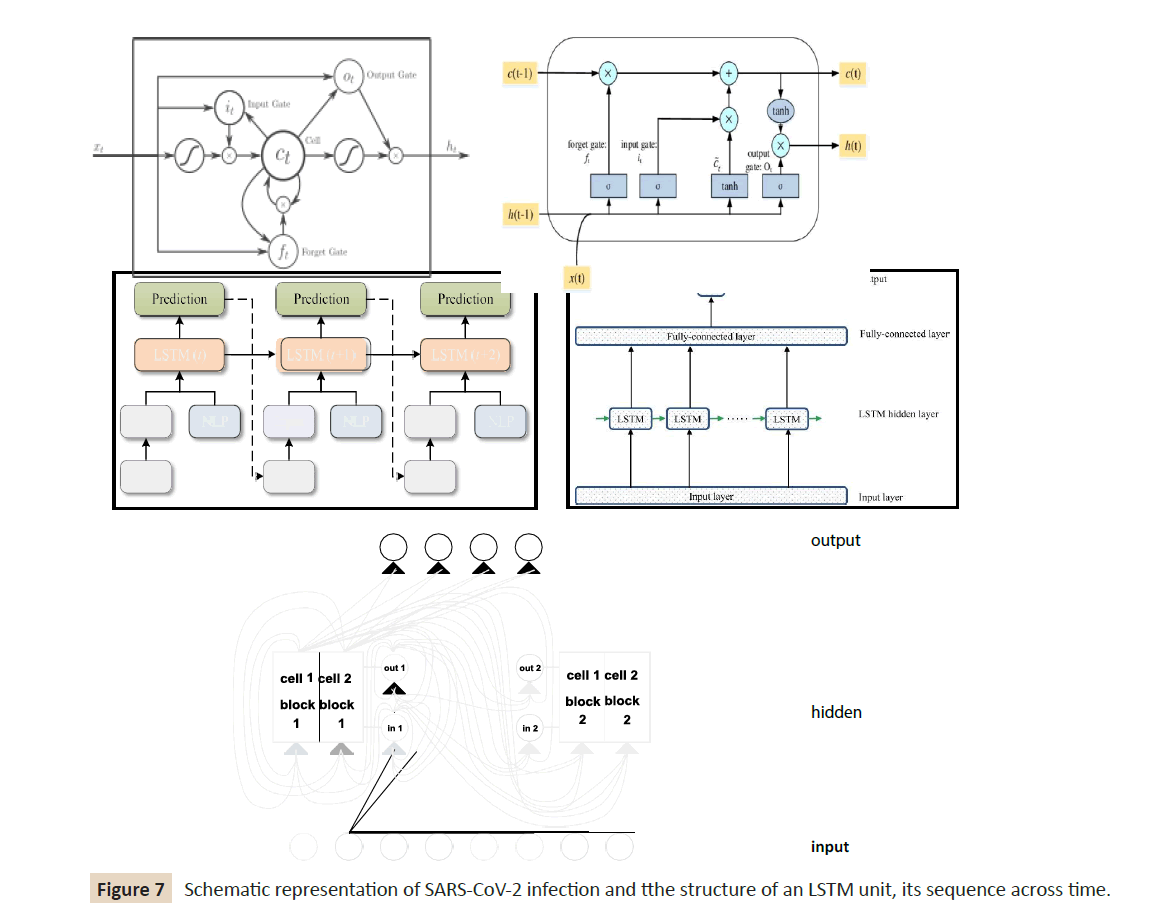
Long short‐term memory networks: An LSTM network is a
subclass of RNNs, trying to circumvent RNNs’ inability to learn
to recognize long-term dependencies in the data sequences
[38,39,60,61]. It is addressed the latter by presenting the LSTM
unit, whereas LSTM networks are constructed by combining
several layers of LSTM units [60]. Specially, (Figure 7) shows the
structure of an LSTM unit and its sequence across time. Every and
each single LSTM unit consists of three gates that operate on the
input vector, xt, to generate the cell state, Ct, and the hidden state,
ht. From a physical interpretation, the cell state can be viewed
as the memory of the cell, while the gates control the flow of
information in and out of the memory. In addition to it, the input
gate determines the incorporation of new information, the forget
gate determines which information should be discarded, and the
output gate controls the in- formation that passes along to the
next layer. Following the interconnections presented in (Figure 7)
the following formulas per category of the variables hold:
Figure 7: Schematic representation of SARS‐CoV‐2 infection and tthe structure of an LSTM unit, its sequence across time.
• Gating variables
ft = σ(Wf xt + Uf ht−1 + bf ) (1)
it = σ(Wixt + Uiht−1 + bi) (2)
ot = σ(Woxt + Uoht−1 + bo) (3)
• Candidate (memory) cell state variable
C t =tanh(Wcxt +Ucht−1 +bc) (4)
• Cell and hidden state variables
Ct=ft ◦ Ct−1+it ◦ C t (5)
ht =ot ◦tanh(Ct) (6)
Where {W, U} and b are the learnable weights and bias of the LSTM
layer, respectively, for the in- put and the recurrent connections
for the input/output/forget gates and cell state; ◦ is the elementwise
product of two vectors; σ is a sigmoid function given by σ (x)
= (1 + e−x) −1 to compute the gate activation function, whereas
the hyperbolic tangent function (tanh) is used to compute the
state activation function [62].
Potential ORF3a protein of SARS-CoV-2 possibility on viral
immune-pathogenicity: Since none of the accessible SARS-CoV-2
genomes are stream-lined in protein databases like STRING
we could not directly get ORF3a (SARS-CoV-2) −human protein
interactome. Our data have significantly established the structural
resemblance of ORF3a protein between SARS-CoV and SARSCoV-
2 thus conceding further functional prediction. Thus, we
deduce the functional pertinence of SARS-CoV-2 putative ORF3a
protein from interactome and pathway enrichment analysis of
SARS-CoV. The aforementioned neutralizing antibody escape
mutations were artificially generated during in vitro replication
of a recombinant virus. However, as monoclonal antibodies
are developed for therapeutic and prophylactic applications, and vaccine candidates are deployed, and the possibility of
SARS-CoV-2 reinfection becomes greater, it is important both
to understand pathways of antibody resistance and to monitor
the prevalence of resistance-conferring mutations in naturally
circulating SARS-CoV-2 populations [63-65]. We used the GISAID
and CoV-Glue, SARS-CoV-2 databases to survey the natural
occurrence of mutations that might confer resistance to the
mono- clonal and plasma antibodies used in our experiments.
Among the SARS-CoV-2 sequences in the CoV2-Glue database at
the time of writing, different non-synonymous mutations were
present in natural populations of SARS-CoV-2 S protein sequences.
Consistent with the finding that none of the mutations that arose
in our selection experiments gave an obvious fitness deficit,
most were also present in natural viral populations. CoV-GLUE
is an online web application for the interpretation and analysis
of SARS- CoV-2 virus genome sequences, with a focus on amino
acid sequence variation [66]. It is based on the GLUE data centric
bioinformatics environment and provides a brows able database
of amino acid replacements and coding region indels that have
been observed in sequences from the pandemic. Users may also
analyse their own SARS-CoV-2 sequences by submitting them to
the web application to receive an interactive report containing visualisations of phylo- genetic classification and highlighting
genomic variation of potentially high impact, for example linked
to primer mismatches (see Table 2 as reference). Position of
neutralization resistance conferring substitutions. Structure of
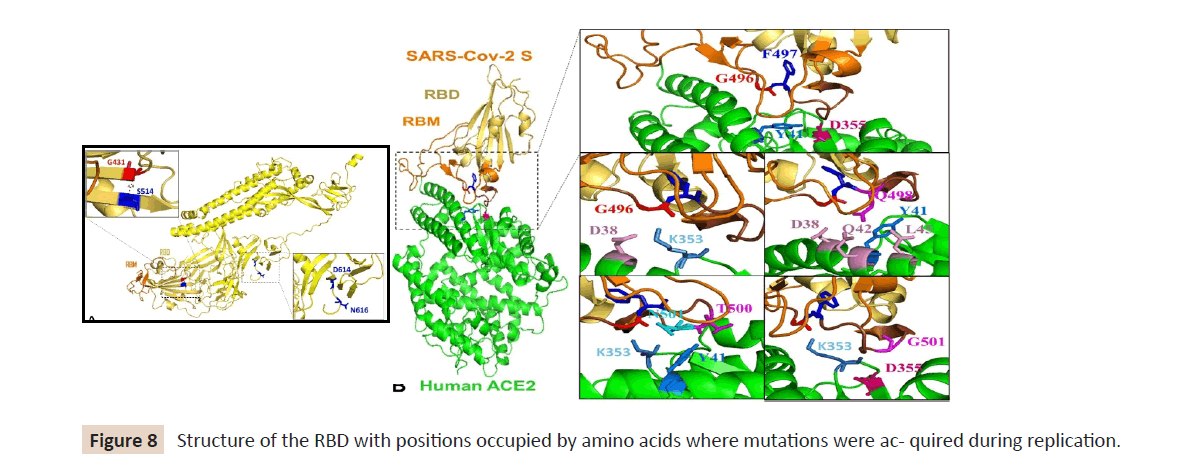
the RBD with positions that are occupied by amino acids where
mutations were acquired during replication in the presence of
each monoclonal antibody indicated (Figure 8). Additionally
(Figure 9) showed the position and frequency of S amino acid
substitutions in SARS-CoV-2 S. We identified and analysed the
amino acid mutations that gained prominence worldwide from
the early months of the pandemic. Eight mutations have been
identified along the viral genome, mostly located in conserved
segments of the structural proteins and showing low variability
among coronavirus, which indicated that they might have a
functional impact. At the moment of writing this paper, these
mutations present a varied success in the SARS-CoV-2 virus
population; ranging from a change in the spike protein that
becomes absolutely prevalent.
Figure 8: Structure of the RBD with positions occupied by amino acids where mutations were ac- quired during replication.
Figure 9: Structural representation of the position and frequency of S amino acid substitutions in SARS-CoV-2 S key residues.
Future selection of combinations of monoclonal
antibodies for therapeutic and prophylactic
applications
The ability of SARS-CoV-2 monoclonal antibodies and plasma to
select variants that are apparently fit and that naturally occur
at low frequencies in circulating viral populations suggests that
therapeutic use of single antibodies might select for escape
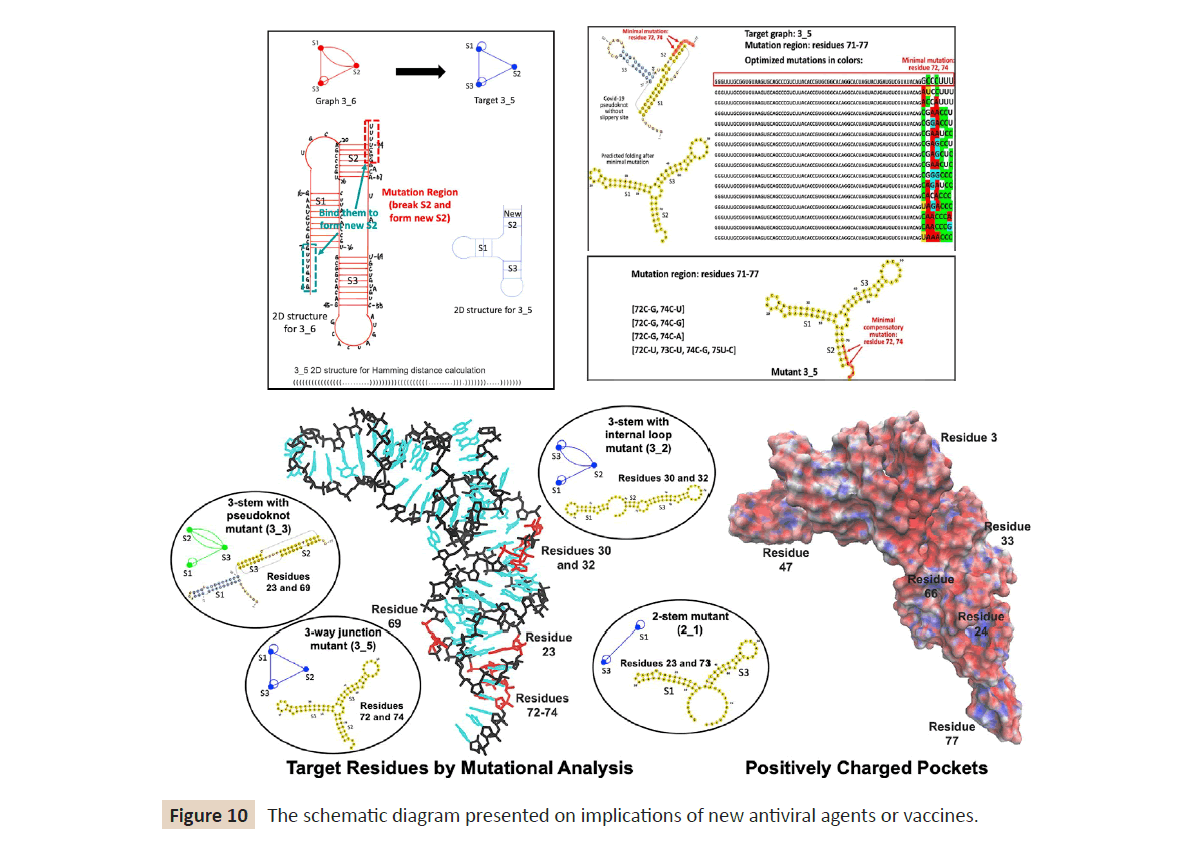
mutants. As shown in (Figure 10) the schematic diagram
presented on implications of new antiviral agents or vaccines. We
tested whether combinations of monoclonal antibodies could
suppress the emergence of resistant variants to mitigate against
the emergence or selection of escape mutations during therapy,
or during population based prophylaxis [67].
Figure 10: The schematic diagram presented on implications of new antiviral agents or vaccines.
Miscellaneous comment on recent spike protein changes: As
seen on many occasions before, mutations are naturally expected
for viruses and are most often simply neutral regional markers
useful for contact tracing. The changes seen have rarely affected
viral fitness and almost never affected clinical outcome but the
detailed effects of these mutations remain to be determined
fully. Changes in the spike protein have relevance for potential
effects on both host receptor as well as antibody binding with
possible consequences for infectivity, transmission potential and
antibody and vaccine escape. Actual effects need to be measured and verified experimentally and GISAID reports updates on
spike mutations of recent submissions via gisaid.org/ spike
and any sequence can be tested for spike mutations via gisaid.
org/covsurver and from the internal analysis interface where
individual countries/regions and time periods can be selected for
custom analysis. This allows highlighting and tracking the rise of
mutations like D614G or the currently most common receptor
binding mutations as well as combinations of these mutations
with deletions altering the spike protein surface [68, 69].
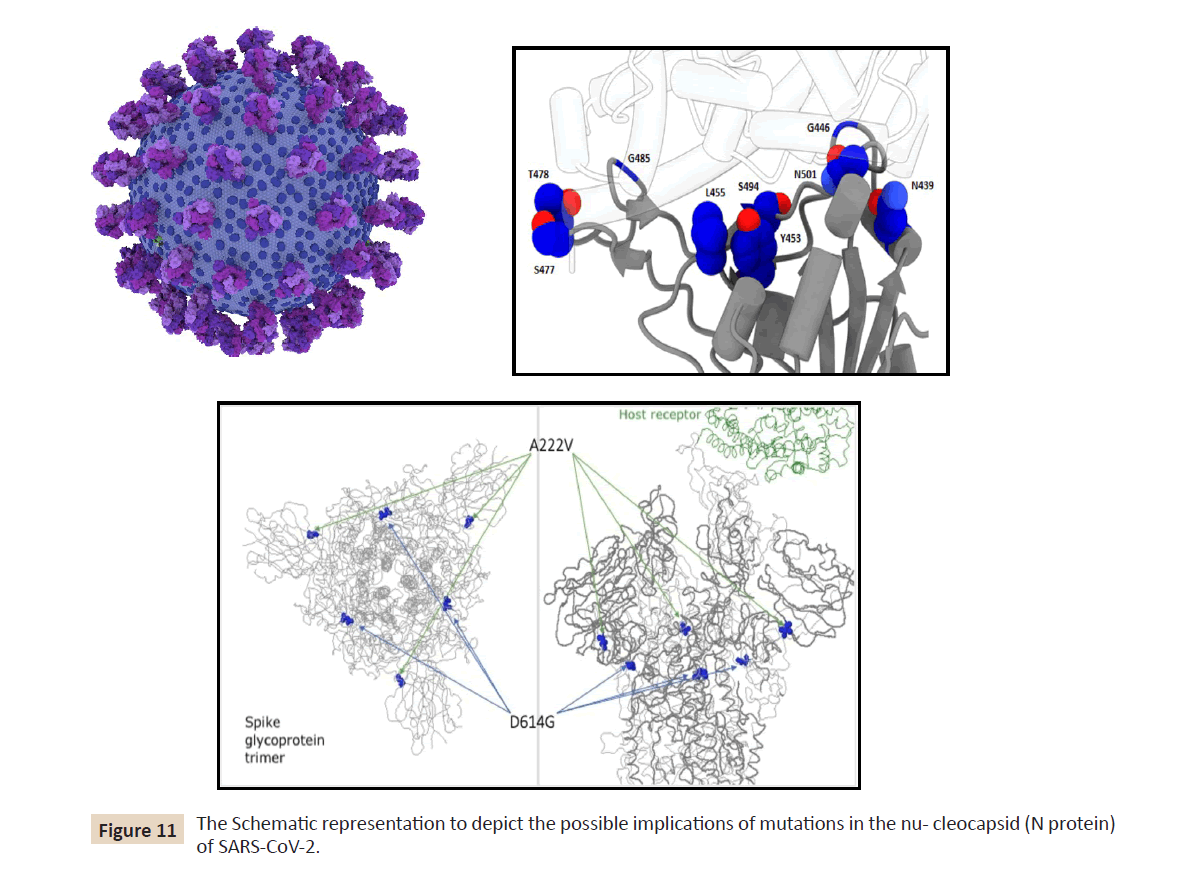
As shown in (Figure 11) it has become evident, these few S gene
mutations and some deletions are found in multiple genomic
contexts (different clades in different countries) that may be an
early indication for some potential advantage for these viruses
but needs to be verified and does not necessarily mean change in
clinical severity or transmission efficiency.
Figure 11: The Schematic representation to depict the possible implications of mutations in the nu- cleocapsid (N protein)
of SARS-CoV-2.
Discussion
The severity of COVID-19 greatly varies from patient to patient.
Majority of the patients either remain asymptomatic or develop
mild to moderate symptoms. However, some COVID-19 patients
who develop severe disease die even after hospitalization and
intensive care. Why the disease severity differs so much from one
person to another is one of the mysteries scientists are still trying
to solve. The present study was designed to explore whether genetic variation in SARS-CoV-2 can explain variable severity of
COVID-19. Mutation profiles of SARS-CoV-2 isolated from mildly
affected and severely affected COVID-19 patients were explored
and compared. Among numerous mutations observed in this
study, two missense mutations, affecting RdRp and spike protein
genes.
Respectively, were found most predominantly in the severely
affected group compared with mildly affected group. Along with
these two mutations in the 5′ UTR and a silent mutation in the
ORF1ab were predominantly found in severely affected group the
later not significantly [70] however, these mutations do not alter
amino acid sequence in a protein. Many other mutations that
were found in low frequency in the present study are unlikely to
exert an effect on the severity of COVID-19. Therefore, the ability
of spike protein and RdRp mutations on the severity of COVID-19
needs to be considered.
Conclusion
Ongoing efforts by the worldwide variants of SARS-CoV-2 are
providing valuable insights into the structural mechanisms of
action between bioinformatics algorithms and predictions.
Structural biology can help explain the effect of amino acid
variations on interactions with other proteins, leading to changes
in the infection rate, associated symptoms and so on. This
knowledge, when combined with structure guided efforts to
design stable vaccine antigens provides an important foundation
for countering the impacts of the disease. These vaccines/drugs
should target the regions of the protein which do not mutate fast.
Therapeutics against regions with high mutation propensities will
make them strain specific. Bioinformatics guided approaches
provide an important framework for understanding the increased
virulence of this pathogen and for designing therapeutics and will
be important for understanding the emergence of drug resistance
and antibody resistant variants/mutations of SARS-CoV-2.
Conflicts of Interest
No potential conflict of interest relevant to this article was
reported.
Acknowledgments
We gratefully acknowledge the authors, originating and
submitting laboratories of the sequences from GenBank and
GISAID’s hCoV- 19 database on which this research is based.
References
- Rodrigues TS, de Sá KSG, Ishimoto AY. (2021) Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med 218: e20201707.
- Pan L, Wang R, Yu N. (2021) Clinical characteristics of re-hospitalized COVID-19 patients with recurrent positive SARS-CoV-2 RNA: a retrospective study [published online ahead of print 2021 Jan 15]. Eur J Clin Microbial Infect Dis 21: 8.
- Baek MS, Cha MJ, Kim MC. (2021) Clinical and radiological findings of adult hospitalized pa-tients with community-acquired pneumonia from SARS-CoV-2 and endemic human coron- aviruses. PLoS One 16: e0245547.
- Choudhary S, Sreenivasulu K, Mitra P, Misra S, Sharma P. (2021) Role of Genetic Variants and Gene Expression in the Susceptibility and Severity of COVID-19. Ann Lab Med 41: 129-138.
- Conti P, Caraffa A, Gallenga CE (2021) The British variant of the new coronavirus-19 (Sars- Cov-2) should not create a vaccine problem [published online ahead of print, 2021 Feb 24]. J Biol Regul Homeost Agents 35: 10.23812/21-3-E.
- Kim C, Ryu DK, Lee J (2021) A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun 12: 288.
- Ahmadpour D, Ahmadpoor P, Rostaing L (2020) Impact of Circulating SARS-CoV-2 Mutant G614 on the COVID-19 Pandemic. Iran J Kidney Dis 14: 331-4.
- Verkhivker GM, Di Paola L. (2021) Dynamic Network Modeling of Allosteric Interactions and Com- munication Pathways in the SARS-CoV-2 Spike Trimer Mutants: Differential Modulation of Conformational Landscapes and Signal Transmission via Cascades of Regulatory Switches. J Phys Chem B 10: 1021/acs.jpcb.0c10637.
- Mittal A, Verma V (2021) Connections between biomechanics and higher infectivity: a tale of the D614G mutation in the SARS-CoV-2 spike protein. Signal Transduct Target Ther 6: 11.
- Gobeil SM, Janowska K, McDowell S. (2021) D614G Mutation Alters SARS-CoV-2 Spike Con- formation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Rep 34: 108630.
- Van Doremalen N, Purushotham J, Schulz J. (2021) Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces shedding of SARS-CoV-2 D614G in rhesus macaques. Preprint bioRxiv.
- Klumpp-Thomas C, Kalish H, Hicks J. (2020) D614G Spike Variant Does Not Alter IgG, IgM, or IgA Spike Seroassay Performance. J Infect Dis jiaa743.
- Lanjanian H, Moazzam-Jazi M, Hedayati M. (2021) SARS-CoV-2 infection susceptibility influ- enced by ACE2 genetic polymorphisms: insights from Tehran Cardio-Metabolic Genetic Study. Sci Rep 11: 1529.
- Rodriguez JH, Gupta A. (2021) Contact residue contributions to interaction energies between SARS- CoV-1 spike proteins and human ACE2 receptors. Sci Rep 11: 1156.
- To KK, Hung IF, Ip JD. (2020) COVID-19 re-infection by a phylogenetically distinct SARS- coronavirus-2 strain confirmed by whole genome sequencing [published online ahead of print, 2020 Aug 25]. Clin Infect Dis ciaa1275.
- Zhao Z, Sokhansanj BA, Malhotra C, Zheng K, Rosen GL. (2020) Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput Biol 16: e1008269.
- Chi X, Yan R, Zhang J. (2020) A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369: 650-5.
- Xia S, Liu M, Wang C. (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high ca- pacity to mediate membrane fusion. Cell Res30:343-55.
- Sedova M, Jaroszewski L, Alisoltani A, Godzik A. (2020) Coronavirus3D: 3D structural visualization of COVID-19 genomic divergence. Bioinformatics 36: 4360-2.
- Wlodawer A, Dauter Z, Shabalin IG. (2020) Ligand-centered assessment of SARS-CoV-2 drug target models in the Protein Data Bank. FEBS J 287: 3703-18.
- Burley SK, Bhikadiya C, Bi C. (2021) RCSB Protein Data Bank: powerful new tools for explor- ing 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res 49: D437-D51.
- Zadeh JN, Steenberg CD, Bois JS. (2011) NUPACK: Analysis and design of nucleic acid sys- tems. J Comput Chem 32:170-3.
- Toubiana D, Puzis R, Sadka A, Blumwald E. (2019) A Genetic Algorithm to Optimize Weighted Gene Co-Expression Network Analysis. J Comput Biol 26: 1349-66.
- Sale M, Sherer EA. (2015) A genetic algorithm based global search strategy for population pharmaco- kinetic/pharmacodynamic model selection. Br J Clin Pharmacol 79: 28-39.
- Kent WJ. (2012) BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656-664.
- Bhagwat M, Young L, Robison RR. Using BLAT to find sequence similarity in closely related genomes. Curr Protoc Bioinformatics Chapter 10: Unit10.8.
- Liu CH, Di YP. (2020) Analysis of RNA Sequencing Data Using CLC Genomics Workbench. Methods Mol Biol 61-113.
- Olotu FA, Omolabi KF, Soliman MES. (2020) Leaving no stone unturned: Allosteric targeting of SARS-CoV-2 spike protein at putative druggable sites disrupts human angiotensin-converting enzyme interactions at the receptor binding domain. Inform Med Unlocked 21: 100451.
- Poosapati A, Gregory E, Borcherds WM, Chemes LB, Daughdrill GW. (2018) Uncoupling the Folding and Binding of an Intrinsically Disordered Protein. J Mol Biol 430: 2389-402.
- Christensen NJ, Kepp KP. (2012) Accurate stabilities of laccase mutants predicted with a modified FoldX protocol. J Chem Inf Model 52:3 028-42.
- Nadra AD, Serrano L, Alibés A. (2011) DNA-binding specificity prediction with FoldX. Methods En- zymol 498: 3-18.
- Kumar R, Jayaraman M, Ramadas K, Chandrasekaran A. (2020) Insight into the structural and func- tional analysis of the impact of missense mutation on cytochrome P450 oxidoreductase. J Mol Graph Model 100: 107708.
- Parthiban V, Gromiha MM, Schomburg D. (2006) CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res 34: W239-W42.
- Gyulkhandanyan A, Rezaie AR, Roumenina L, Lagarde N, Fremeaux-Bacchi V, et al. (2020) BO. Analysis of protein missense alterations by combining sequence- and structure- based methods. Mol Genet Genomic Med 8: e1166.
- Wu K, Wei GW. (2018) Quantitative Toxicity Prediction Using Topology Based Multitask Deep Neural Networks. J Chem Inf Model 58: 520-31.
- Chen J, Wang R, Wang M, Wei GW. (2020) Mutations Strengthened SARS-CoV-2 Infectivity. J Mol Biol 432: 5212-26.
- Cheng MH, Zhang S, Porritt RA, Arditi M, Bahar I. (1997) An insertion unique to SARS-CoV-2 ex- hibits superantigenic character strengthened by recent mutations. Preprint. bioRxiv 9:1735-80.
- Liu X, Liu C, Huang R. (2020) Long short-term memory recurrent neural network for pharmaco- kinetic-pharmacodynamic modeling. Int J Clin Pharmacol Ther 10.5414/CP203800.
- Maragatham G, Devi S. (2019) LSTM Model for Prediction of Heart Failure in Big Data. J Med Syst 43(5): 111.
- Eskier D, Suner A, Karakülah G, Oktay Y. (2020) Mutation density changes in SARS-CoV-2 are relat- ed to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ 8: e9703.
- Brown J, Pirrung M, McCue LA. (2017) FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33: 3137-39.
- Chen H, Li Y, Sun W, Song L, Zuo R, et al. (2020) Characterization and source identification of an- tibiotic resistance genes in the sediments of an interconnected river-lake system. Environ Int 137: 105538.
- Hou YJ, Chiba S, Halfmann P. (2020) SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370: 1464-8.
- Plante JA, Liu Y, Liu J. (2020) Spike mutation D614G alters SARS-CoV-2 fitness and neutraliza- tion susceptibility. bioRxiv 09.01.278689.
- Zhang L, Jackson CB, Mou H. (2020) The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. Preprint. bioRxiv 06.12.148726.
- Korber B, Fischer WM, Gnanakaran S. (2020) Tracking Changes in SARS-CoV-2 Spike: Evi- dence that D614G Increases Infectivity of the COVID-19 Virus Cell 182: 812-27 e19.
- Grubaugh ND, Hanage WP, Rasmussen AL. (2020) Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell 182: 794-5.
- Weissman D, Alameh MG, de Silva T. 29: 23-31 e4.
- Forni D, Cagliani R, Pontremoli C. (2020) Antigenic variation of SARS-CoV-2 in response to immune pressure [published online ahead of print, 2020 Dec 2]. Mol Ecol 10.1111/mec 15730.
- Yurkovetskiy L, Wang X, Pascal KE. (2020) Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 183:739-51 e8.
- Poran A, Harjanto D, Malloy M. (2020) Sequence-based prediction of SARS-CoV-2 vaccine tar- gets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med 12: 70.
- Kosuge M, Furusawa-Nishii E, Ito K, Saito Y, Ogasawara K. (2020) Point mutation bias in SARS- CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci Rep 10: 17766.
- Pachetti M, Marini B, Benedetti F. (2020) Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med 18: 179.
- Lau SY, Wang P, Mok BW. (2020) Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect. 9: 837-42.
- Daniloski Z, Guo X, Sanjana NE. (2020) The D614G mutation in SARS-CoV-2 Spike increases trans- duction of multiple human cell types. Preprint bioRxiv.
- Li Q, Wu J, Nie J. (2020) The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 182:1284-94 e9.
- Shajahan A, Supekar NT, Gleinich AS, Azadi P. (2020) Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 30: 981-8.
- Allen JD, Watanabe Y, Chawla H, Newby ML, Crispin M. (2020) Subtle Influence of ACE2 Glycan Processing on SARS-CoV-2 Recognition [published online ahead of print, 2020 Dec 17]. J Mol Biol 433: 166762.
- Pujić I, Perreault H. (2021) Recent advancements in glycoproteomic studies: Glycopeptide enrichment and derivatization, characterization of glycosylation in SARS CoV2, and interacting glycopro- teins. Mass Spectrom Rev 10.1002
- Hochreiter S, Schmidhuber J. (1997) Long short-term memory. Neural Comput 9: 1735-80.
- Gers FA, Schmidhuber J. (2000) Cummins F. Learning to forget: continual prediction with LSTM. Neural Comput 12: 2451-71.
- Guo H, Sung Y. (2020) Movement Estimation Using Soft Sensors Based on Bi-LSTM and Two-Layer LSTM for Human Motion Capture. Sensors (Basel) 20: 1801.
- Oberemok VV, Laikova KV, Yurchenko KA, Fomochkina II, Kubyshkin AV. (2020) SARS-CoV-2 will continue to circulate in the human population: an opinion from the point of view of the virus- host relationship. Inflamm Res 69: 635-40.
- Weisblum Y, Schmidt F, Zhang F. (2020) Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9: e61312.
- van Dorp L, Richard D, Tan CCS, Shaw LP, Acman M. (2020) evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun 11: 5986.
- Singer J, Gifford R, Cotton M, Robertson D. (2020) CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation.
- Rilinger J, Kern WV, Duerschmied D. (2020) A prospective, randomised, double blind placebo- controlled trial to evaluate the efficacy and safety of tocilizumab in patients with severe COVID-19 pneumonia (TOC-COVID): A structured summary of a study protocol for a randomised controlled trial. Trials 21: 470.
- Ortega JT, Serrano ML, Pujol FH, Rangel HR. (2020) Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J 19: 410-7.
- Hussain M, Jabeen N, Raza F. (2020) Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol 92: 1580-6.
- Baldassarre A, Paolini A, Bruno SP, Felli C, Tozzi AE, et al. (2020) Potential use of noncoding RNAs and innovative therapeutic strategies to target the 5'UTR of SARS-CoV-2. Epigenomics 12:1349-61.