Keywords
Bronchial asthma; Interleukin-4; IgE; STAT6; Gene polymorphism
Introduction
Bronchial asthma is the most common chronic disease among children in the world. It affects up to 300 million people worldwide [1]. The prevalence among Egyptian children aged 3–15 years was 8.2% [2]. Bronchial asthma is influenced by genetic and environmental factors. The disease is defined by the presence of airway hyperreactivity, mucus overproduction, and chronic eosinophilic inflammation [3].
Asthma is often characterized by enhanced total serum IgE level upon the exposure to allergens, which is known as an atopy. Many family studies, through genome-wide linkage studies, confirmed the involvement of genetic predisposition in the development of atopy in asthmatic patients [4]. The elevated IgE production in asthmatic patients results in promotion of acute hypersensitivity responses, chronic eosinophil-predominant allergic inflammation with T helper-2 (Th2) cells cytokine production [5].
IL-4 serves as an essential proinflammatory cytokine in immune regulation mediated by activated T helper cells (Th) and facilitates immunoglobulin E isotype switching in B cells, growth, and differentiation of B cells and monocytes. As an important signal molecule, IL-4 can exacerbate airway inflammation through modulating eosinophils, lymphocytes, and air epithelial cells that play an important role in the pathogenesis of asthma. Moreover, IL-4 plays a pivotal role in phenotypic changes of bronchial asthma, such as airway hyperresponsiveness, eosinophil infiltration, and mucus overproduction [6]. Also STAT6, the signaling molecule from JAK/STAT pathway, activated by IL-4 and IL-13 cytokines, plays an important role in IgE production and allergic airway inflammation [5].
Signal transducer and activator of transcription (STAT) is a family of latent cytoplasmic transcription factors activated by specific cytokine receptor-mediated signal transducers. Selective activation of STAT6 by IL-4 or IL-13 involves phosphorylation, dimerization and then translocation into the nucleus, where it binds to specific DNA elements TTC (N3/4) GAA within the promoter region, activating gene transcription [7].
STAT6 gene, 19 kb with 23 exons and located on chromosome 12q13.3–q14.1, is highly polymorphic and thousands of SNPs within both coding and non-coding regions have been collected in the dbSNP database. STAT6 rs324011 (2892C/T) SNP is found in the second intron; interestingly, the C→T substitution creates an additional putative binding site for Nuclear Factor κB (NF-κB), therefore, enhancing NF-κB-mediated STAT6 gene transcription with concomitant elevated STAT6-mediated IgE production [8].
Therefore, the present study was undertaken to investigate the relationship between STAT6 gene polymorphism and the susceptibility to atopic asthma in Egyptian children and to correlate the effect of STAT6 gene polymorphism on IL-4 and total IgE levels.
Methods
Study groups
The study included 90 Egyptian children ; Sixty children with atopic asthma diagnosed according to the criteria of the Global Initiative for Asthma (GINA), who were presented to the Outpatient Clinic of the Pediatric Department, Tanta University Hospital in the period from June 2015 to March 2016 and 30 healthy children with matched as a control group. All asthma patients were clinically stable. None of the participants received antihistamine, systemic or topical corticosteroids in 3 weeks prior to clinical evaluation. Subjects excluded from this study were those with heart failure, renal failure, liver failure, diabetes mellitus, malignancy and tuberculosis. Ethical approval of this study was obtained from the Ethical Committee of Faculty of Medicine Tanta University (Approval code 2820/10/14) and written informed consents were obtained from all children parents.
All participants underwent a full clinical examination, skin prick testing, routine and specific laboratory investigations.
Total immunoglobulin E (IgE) and Prick test assays
Atopy was diagnosed by a positive skin prick test (wheal diameter ≥3 mm) to at least one of 12 common aeroallergens and by measurement of the total IgE level. Positive values were taken to be ≥200 IU/ml. The control subjects had no history of asthma or other allergic diseases, negative skin prick tests and normal total IgE values.
Blood sample collection
Five milliliter venous blood were taken from every investigated subject under complete aseptic conditions and divided into 2 portions: one mL of whole blood was collected in sterile ethylenediaminetetraacetic acid (EDTA)-containing tubes for DNA extraction and eosinophil counts and stored at – 80°C for further PCR amplification, and the remainder was left for 30 to 60 minutes for spontaneous clotting at room temperature before being centrifuged at 3000 rpm for 10 minutes. Serum samples were kept frozen at –20°C for determination of total IgE and IL-4 levels. Eosinophil counts were determined according to Burrows et al. [9].
Assay of serum total IgE and IL-4 levels
Serum total IgE was measured using Immunospec® human IgE sandwich enzyme linked immunosorbent assay (ELISA) kit (Cat# 10602, Vivantis International, Egypt). Serum IL-4 was estimated using Bioseps® human interleukin-4 ELISA kit (Cat# BEK1103, Vivantis International, Egypt). Both assays were carried out according to the manufacturers’ instructions. Using the mean absorbance value for each sample, the corresponding concentration of IgE and IL-4 were determined from the standard curve.
Genomic DNA extraction
Genomic DNA was extracted from EDTA whole blood using a spin column method according to the manufacturer’s instructions. (Cat# GF-BT-100, Vivantis Inc. USA). The quality of DNA was assessed by measuring absorbance at both 260 nm and at 280 nm. DNA was stored at − 20 °C till the time of use.
Genotyping for the STAT6 rs324011 polymorphism
Polymerase Chain Reaction-restriction fragment length polymorphism (PCR-RFLP) was performed using the thermal cycler (Whatman Biometra, Germany) to determine the different genotypes of STAT6-2892C/T. The specific primers were: Fwd: 5'-CTCTTCCCACCCCTGTGTCTATC3'; and Rev: 5'- TCCCATAGATAGCCCTCCTAGGTAC-3' [10]. Briefly, the PCR mixture (total, 25 μl) containing 5 μL DNA, 3 μl of each primer, 12.5 μL 2X Taq master mix (containing Taq DNA Polymerase, dNTPs, MgCl2 and reaction buffers at optimal concentrations for efficient amplification of DNA templates by PCR) and 1.5 μL nuclease free water. The PCR mixture was incubated at 95?C for 5 minutes, then 35 cycles of 95?C for 30 seconds minute to denature, 72?C for 40 seconds to anneal the primers and 72?C for 8 minute to elongate the strand PCR, amplification kit was obtained from ((SIGMA Chemical Co, St. Louis, Missouri, USA).
Subsequently, the amplified products were digested by the BshNI (Cat# RE1106, Vivantis Inc. USA) restriction enzyme according to the manufacturer’s instructions. Digested products were separated on 3% agarose gel stained with ethidium bromide. The gel was visualized under a UV transilluminator with a 10 bp marker ladder (Ultra Low Range DNA Ladder) (34-501 bp) served as reference for DNA fragment size (Cat# NM2415,Vivantis Inc. CA, USA) and photographed. The STAT6-2892C/T alleles were differentiated by BshNI restriction digestion (C allele, a single fragment of 132 bp; T allele, fragments of 107 and 25bp (obscure band)).
Statistical Analysis
The results for continuous variables were expressed as means ± SD and t-test and one-way analysis of variance (ANOVA) were used for their analysis. Genotype frequencies of STAT6 genotypes in atopic asthmatic children and controls were tested for Hardy–Weinberg equilibrium, and any deviation between the observed and expected frequencies was tested for significance using the Chi-squared test. The statistical significances of differences in frequencies of variants between the groups were tested using the χ2 test. In addition, the odds ratios (OR) and 95% confidence intervals (CIs) were calculated as a measure of the association of the polymorphic genotypes (CT and TT) with atopic asthma susceptibility. A difference was considered significant at P-values less than 0.05. All statistical calculations were performed using the computer program SPSS (Statistical Package for the Social Science) version 17 for Microsoft Windows.
Results
Demographic and biochemical data of the two studied groups is illustrated Table 1. There were no statistically significant differences among the two groups as regard age, sex distribution or exposure to passive smoking (p>0.05).
| Variables |
Healthy control subjects (n=30) |
Atopic asthmatic children (n=60) |
Statistical test |
| t/ X2 |
P-value |
| Age |
9.30 ± 1.82 |
8.90 ± 1.68 |
t= 1.03 |
>0.05 |
| Sex (Males/Females) |
11/19 |
26/34 |
X2=0.37 |
>0.05 |
| Duration of Asthma (years) |
NA |
5.9 ± 1.5 |
- |
- |
| Passive smoking% |
| Negative |
0.6 |
0.54 |
X2=0.73 |
>0.05 |
| Positive |
0.4 |
0.46 |
| Total IgE (IU/ml) |
25.25 ± 19.21 |
670.08 ± 136.56 |
t= 25.67 |
<0.0001* |
| Eosinophils % |
1.43 ± 0.74 |
8.53 ± 1.63 |
t= 22.67 |
<0.0001* |
Data are presented as mean ± SD or percentages. P was calculated by Chi-square or unpaired t -test. P was considered significant at <0.05.
Table 1: Demographic and biochemical data of healthy control subjects and atopic asthmatic children.
The disease duration of the asthmatic group was 5.9 ± 1.5 years. Total serum IgE and peripheral blood esinophils % were significantly higher in atopic asthmatic children as compared to control subjects (p<0.0001). Also Interlukin-4 serum levels were significantly increased in atopic asthmatic children as compared to the healthy control subjects (p<0.0001) as depicted in Figure 1.
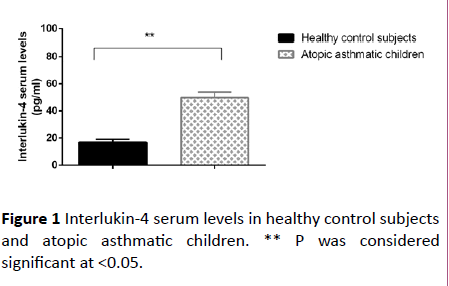
Figure 1: Interlukin-4 serum levels in healthy control subjects and atopic asthmatic children. ** P was considered significant at <0.05.
As regard the Genotypes of the STAT6-2892C/T polymorphism in the present study, three genotypes for STAT6-2892C/T polymorphism were recognized by genotyping; (CC) homozygous CC genotype with one band at 132 bp due to absence of the BshNI restriction site; homozygous TT genotype with 2 bands at 25 bp and 107 bp due to presence of the BshNI restriction site; and heterozygous CT genotype with 3bands at 25 bp, 107 bp, and 132 bp (Figure 2).
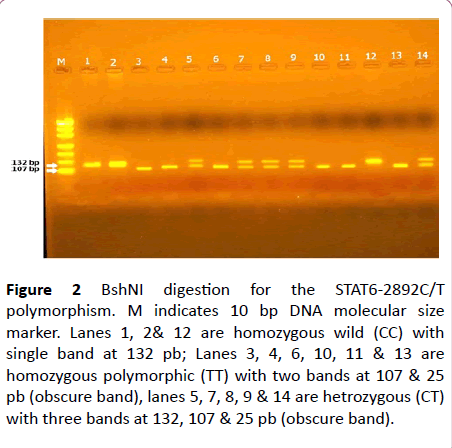
Figure 2: BshNI digestion for the STAT6-2892C/T polymorphism. M indicates 10 bp DNA molecular size marker. Lanes 1, 2& 12 are homozygous wild (CC) with single band at 132 pb; Lanes 3, 4, 6, 10, 11 & 13 are homozygous polymorphic (TT) with two bands at 107 & 25 pb (obscure band), lanes 5, 7, 8, 9 & 14 are hetrozygous (CT) with three bands at 132, 107 & 25 pb (obscure band).
The distribution of genotypes of STAT6-2892C/T polymorphism in controls were consistent with expectations under the Hardy–Weinberg equilibrium (X2 = 2.93, p= 0.089). In contrast, the observed genotype distributions deviated from those predicted by the Hardy–Weinberg equilibrium in atopic asthmatic children (P<0.05) as shown in Table 2.
| Groups/Genotypes |
Observed frequency |
Expected frequency |
X2 |
P-value |
| CC |
CT |
TT |
CC |
CT |
TT |
| Healthy Control Subjects |
20 |
7 |
3 |
18.4 |
10.2 |
1.4 |
2.93 |
>0.05 |
| Atopic Asthmatic Children |
24 |
16 |
20 |
17.05 |
29.87 |
13.08 |
10.86 |
<0.05 |
Table 2: Hardy Weinberg equilibrium for STAT6 (2892 C/T) gene polymorphisms in healthy control subjects and atopic asthmatic children.
The genotypic and allelic frequencies of STAT6-2892C/T gene polymorphisms in healthy control subjects and atopic asthmatic children are demonstrated in Table 3. The CC genotype and the C allele were taken as references. The distribution of the STAT6-2892C/T genotypes and alleles differed significantly between atopic asthmatic children and healthy control subjects (X2 = 7.25, 10.58) respectively, p<0.05.
| Geno-typing |
Healthy control subjects (n=30) |
Atopic asthmatic children (n=60) |
OR 95% CI |
P value |
Chi-Square |
| No. |
Frequency |
No. |
Frequency |
X2 |
P-value |
| CC |
20 |
66.70% |
24 |
40% |
1 |
- |
|
| Reference |
| CT |
7 |
23.30% |
16 |
26.70% |
1.9 |
0.65-5.54 |
0.23 |
7.25 |
0.02* |
| TT |
3 |
10.00% |
20 |
33.30% |
5.55 |
1.43-21.44 |
0.01* |
|
|
| C allele |
47 |
78.33% |
64 |
53.30% |
1 |
- |
10.58 |
0.001* |
| Reference |
| T allele |
13 |
21.67% |
56 |
46.70% |
3.16 |
1.55-6.44 |
0.001* |
* P was considered significant at <0.05. OR: odds ratio; CI: confidence interval.
Table 3: Genotype and allele frequencies of STAT6 (2892 C/T) gene polymorphism in healthy control subjects and atopic asthmatic children.
The frequencies of the CT genotype were insignificantly higher in atopic asthmatic children (26.7%) than healthy control subjects (23.3%), where the odds ratios were 1.90 (95% CI: 0.65 -5.54), p>0.05. However, TT genotype frequency was significantly higher in atopic asthmatic children (33.3%) than healthy control subjects (10%), where its odds ratios were 5.55 (95% CI: 1.43-21.44), p<0.05. Meanwhile, the frequency of T allele was significantly higher in atopic asthmatic children (46.7%) than healthy control subjects (21.67%), where odds ratios were 3.16 (95% CI: 1.55-6.44), p<0.05. For further assessment the association of different STAT6-2892C/T genotypes with serum IL-4 levels, atopic asthmatic children were subdivided according to C/T genotype where higher serum IL-4 levels were significantly associated with the minor T allele carriers (CT and TT genotypes) compared to the CC genotype, p<0.0001.
Moreover, the atopic asthmatic children with TT genotype had significantly higher IL-4 serum levels as compared to those with CT genotype, p<0.0001. Likewise, total serum IgE and peripheral blood esinophils% were significantly higher in atopic asthmatic children with CT and TT genotypes compared to the CC genotype (p<0.01) with being significantly higher in atopic asthmatic children with TT as compared to those with CT genotype, p<0.01. These data are summarized in Table 4.
| Parameters |
STAT6 (2892 C/T) genotypes |
One way ANOVA |
CC
(n=24) |
CT
(n=16) |
TT
(n=20) |
F |
P-value |
| IL-4 (pg/ml) |
45.97 ± 2.19 |
49.55 ± 1.91# |
52.67 ± 3.59# |
34.28 |
<0.0001* |
| Total IgE (IU/ml) |
576.70 ± 41.31 |
672.45 ± 74.11# |
735.77 ± 164.15# |
12.71 |
<0.0001* |
| Eosinophils % |
8.00 ± 1.33 |
7.50 ± 2.16 |
9.35 ± 1.21# ¶ |
7.08 |
0.0018* |
Data presented as means ± SD, *P was considered significant at <0.05, # significane Vs CC genotype, ¶ significane Vs CT genotype
Table 4: Association between the distribution of STAT6 (2892 C/T) genotypes and the studied parameters in Atopic asthmatic children.
Discussion
Asthma is one of the most common chronic diseases in childhood. For a long time, asthma has been known to cluster in families, and family studies were the first to suggest that the disease was genetically inherited [11]. One of the most replicated asthma candidate gene is signal transducer and activator of transcription 6 (STAT6), a regulator of IgE class switching. STAT6 may promote the development of asthma by increasing serum IgE level and facilitating airway hyperresponsivness [12]. So the present study aimed to investigate whether single nucleotide polymorphism (SNP) in STAT6 is associated with atopic asthma susceptibility in Egyptian children and to correlate the effect of STAT6 gene polymorphism on IL-4 and total IgE levels.
Our data pointed out that the TT variant genotype of the STAT6 polymorphism rs324011 were significantly associated with an increased risk of atopic bronchial asthma with reference to the CC and CT genotypes, indicating a potentially crucial role for this gene in the pathogenesis of asthma. These data are consistent with the findings of Wu et al. [10] who reported that STAT6 rs324011 may increase an individual’s susceptibility to atopic asthma and contribute to the pathogenesis of asthma in middle China. Li et al. [13] performed a meta-analysis for evaluation of the association between rs324011 polymorphism in the STAT6 gene and the risk of bronchial asthma and their results draw the conclusion that the association between STAT6 rs324011 polymorphism and bronchial asthma was definite and determined. They also documented that rs324011 homozygous TT genotype contributed to the etiology or susceptibility to asthma. Moreover, Al-Muhsen et al. [14] found a significant association of rs324011 homozygous TT genotype with the risk of atopic bronchial asthma whereas rs324015 genotypes were not in the Saudi population. Well in line, haplotype analysis of Godava et al. [15] revealed that rs324011, rs3024974 and rs4559 polymorphisms in STAT6 were associated with total IgE elevation and increased risk of atopic bronchial asthma. STAT6 intron 2 acts as a silencer regulatory element. The polymorphic T allele at rs324011 increases STAT6 promoter activity and gene expression of STAT6 compared with the wild-type C allele. These effects correlate with the creation of a novel, T-allele-specific binding site for the transcription factor NF-κB in T cells [8].
However, some previous findings are inconsistent with our results, Pykalainen et al. [16] documented no signi?cant association of rs324011 to asthma or serum total IgE levels in the Finnish population. Moreover, the study Kavalar et al. [12] and Berenguer et al. [17] did not support such an association between the STAT6 rs324011 and the risk of atopic bronchial asthma in Slovenian children and the Madeira island population, respectively.
The diversity of these studies has pointed out that environmental influences and epigenetic mechanisms as methylation and histone acetylation could pose important modifying factors [18]. Furthermore, gene-gene and gene-environment interactions are involved in the pathogenesis of asthma and more often present in patients with asthma or those with increased total IgE levels [19].
Moreover, the current study focused on the roles of IL-4 in the pathogenesis of bronchial asthma in children. Our results confirmed significantly higher serum IL-4 levels in atopic asthmatic children as compared to controls. Supporting our findings, Bao et al. [20], Shi et al. [21], Wu et al, [22] and Nesi et al. [23] revealed a significant increase in IL-4 in ovalbumin-induced asthmatic mice compared to the control. Our finding may also be reinforced in the light of observations of Antczak et al. [5] who found higher mRNA level of IL-4 and IL-13 genes in asthmatic patients when compared to controls. But they did not find statistically significant differences between controlled and uncontrolled bronchial asthma, even the highest IL-4 expression was recognized in uncontrolled atopic asthma patients. This observation may suggest that IL-4 expression may play a significant role in the course of asthma but it is not a sufficient inducer for asthma.
Interleukin-4 can exacerbate airway in?ammation through modulating eosinophils, lymphocytes, and air epithelial cells that play an important role in the pathogenesis of bronchial asthma. Also, IL-4 is reported to play a pivotal role in phenotypic or functional changes of bronchial asthma, such as airway hyperresponsiveness, eosinophil in?ltration, and mucus overproduction [24].
Emerging evidences have shown that IL-4 messenger ribonucleic acid and protein were highly expressed in airway mucosa of asthmatic patients; abnormal increase of IgE, promoted by IL-4, has been proved to be one of the pathogenesis of asthma, which indicates that IL-4 may induce asthma indirectly [25]. It has been revealed that, IL-4 is signi?cantly overexpressed during the acute and plateau stage of asthma, indicating that immune dysfunction participates in the development and progression of asthma [26].
Immunoglobulin E is a key downstream biomarker of Th2 in?ammation. IgE binds to the high-af?nity IgE receptor FcεRI on mast cells and basophils, and antigen cross linking of IgE leads to degranulation and release of in?ammatory mediators, including histamine, prostaglandins, and pro-in?ammatory cytokines (IL-4, IL-5, IL-13). In the lower airways, this activity may result in eosinophilia, increased mucus production, and enhanced smooth muscle contractility [27].
In the present study, total plasma IgE levels were found to be significantly higher in atopic asthmatic children as compared to controls. Our findings are in accordance with the results of Ghadah [28], Noureldin et al. [29] and Alasandagutti et al. [30] who reported higher total IgE levels in atopic asthmatic group than normal controls. It has been revealed that the production of IgE from B cells and the differentiation, maturation, migration and survival of eosinophils are induced by the increased pro-inflammatory cytokines, including interleukins IL-4, IL-5 and IL-13 [31].
Many types of cells are involved in the pathophysiology of asthma. The contribution of mast cells, lymphocytes, and eosinophils in the induction and effector phase has been well established [32]. Interestingly, eosinophils are the ?rst cells recruited to the site of the allergic reaction and they play important roles in the pathogenesis of asthma in the way of releasing inflammatory mediators, including radical oxygen species, cysteinyl leukotrienes (CysLTs), major basic protein (MBP) and cytokines, that linked the presence of eosinophils to the persistence of inflammatory infiltrate, tissue damage and remodeling [33].
In the current study, as regards peripheral blood eosinophils, they were found to be significantly higher in atopic asthmatic children as compared to controls. In agreement with our findings Ma et al. [34], Wu et al. [22] and Malmström et al. [35] who found significantly higher peripheral eosinophil counts in asthmatic group. Also, Chen et al. [27] reported that there were statistically significant increase in the numbers of total leukocytes, lymphocytes, monocytes, eosinophils and basophils in the asthmatic group in comparison to control one.
Interestingly, IL-4 prolonges the survival of lung effector memory T cells, enhances Th2 cytokine production and also increases the expression of chemokine ligand 11 (CCL11), IL-5, IL-9, and IL-13 in ILC2 [36]. These Th2 cytokines and chemokines may attract eosinophils into the airway lumen and prolong survival of basophils and eosinophils. In addition, the upregulation of vascular cell adhesion molecule-1 (VCAM-1) via IL-4 promotes eosinophil recruitment to the target tissues [37].
Additionally, we revealed statistically significant positive correlations between the rs324011 TT and CT genotypes, and total level of serum IgE. It could indicate the role of this polymorphism in atopic bronchial asthma pathogenesis. Although the single polymorphism rs324011 is located in the second intron of the STAT6 gene, it may have a functional importance. The second Intron was identified as a cis-acting silencing element leading to a reduction of STAT6 promoter activity. In vitro, promoter activity is dependent on the allelic state of rs324011 [8] confirmed that the T allele of rs324011 increases STAT6 promoter activity by creating a new site for NF-κB binding which is not detected in the wild C allele.
Furthermore, we also found significant positive correlation between rs324011 TT and CT genotypes and serum IL-4 levels. In agreement with our findings, Gene expression analysis of Li et al. [38] showed that patients with 4 SNPs (rs2243250, rs1800925, rs1805010, and rs3224011) in STAT6 gene had higher expression levels of IL-4, IL-13, and STAT6. It has been also detected that in bronchial asthma the over expression of IL-4 occurs via the activation of the STAT6 pathway thus showing that STAT6 plays a positive role, affecting the expression of IL-4 [39].
Our study has some limitations that worth mentioning. First, the sample size is relatively small; therefore, our findings should be interpreted with caution and verified using larger scaled studies. Also, because this study is restricted to the Egyptian population; these results may not be applied to the patients of other ethnic or racial groups.
Conclusion
Our study demonstrated that STAT6-2892C/T polymorphism contributes significantly to IgE production and the susceptibility of atopic asthma in Egyptian children, and highlights the etiological relationship between IL-4/STAT6 signaling pathway and atopic bronchial asthma. Given that STAT6 gene does not work alone as it cooperates with other genes in IL-4 /IL-13 signaling pathway, therefore the study of gene-gene and gene-environment interactions is greatly warranted in the future.
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- Tsabouri S, Mavroudi A, Feketea G, Guibas GV (2017) Subcutaneous and sublingual immunotherapy in allergic asthma in children. Front Pediatr 5: 82-97.
- Abd El-Salam M, Hegazy AA, Adawy ZR, Hussein NR (2014) Serum level of naphthalene and 1,2benz-anthracene and their e?ect on the immunologic markers of asthma and asthma severity in children-Egypt. Public Health Res 4: 166-172.
- Liu Y, Zhang H, Ni R, Jia WQ, Wang YY (2017) IL-4R suppresses airway inflammation in bronchial asthma by inhibiting the IL-4/STAT6 pathway. Pulm Pharmacol Ther 43: 32-38.
- Ober C, Yao TC (2011) The genetics of asthma and allergic disease: A 21st century perspective. Immunol Rev 242: 10-30.
- Antczak A, Doma?ska-Senderowska D, Górski P, Pastuszak-Lewandoska D, Nielepkowicz-Go?dzi?ska A, et al. ( 2016) Analysis of changes in expression of IL-4/IL-13/STAT6 pathway and correlation with the selected clinical parameters in patients with atopic asthma. Int J Immunopathol Pharmacol 29: 195-204.
- Wynn TA (2015) Type 2 cytokines: Mechanisms and therapeutic strategies. Nat Rev Immunol 15: 271-282.
- Wang C, Zhu C, Wei F, Gao S, Zhang L, et al. (2017) Nuclear localization and cleavage of STAT6 is induced by kaposi’s sarcoma-associated herpesvirus for viral latency. PLoS Pathog 13: e1006124
- Schedel M, Frei R, Bieli C, Cameron L, Adamski J, et al. (2010) An IgE-associated polymorphism in STAT6 alters NF-kB binding, STAT6 promoter activity, and mRNA expression. J Allergy Clin Immunol 124: 583-589.
- Burrows B, Hasan FM, Barbee RA, Halonen M, Lebowitz MD (1980) Epidemiologic observations on eosinophilia and its relation to respiratory disorders. Am Rev Respir Dis 122: 709-719
- Wu X, Li Y, Chen Q, Chen F, Cai P, et al. (2010) Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle china. J Asthma 47: 238-244.
- Shaker OG, Sadik NA, El-Hamid NE (2013) Impact of single nucleotide polymorphism in tumor necrosis factor-α gene 308G/A in Egyptian asthmatic children and wheezing infants. Human Immunol 74: 796-802.
- Kavalar MS, Balantic M, Silar M, Kosnik M, Korosec P et al. Association of ORMDL3, STAT6 and TBXA2R gene polymorphisms with asthma. Int J Immunogenet 39: 20-25.
- Li B, Nie W, Li Q, Liu H, Liu S (2013) Signal transducer and activator of transcription 6 polymorphism and asthma risk: A meta-analysis. Int J Clin Exp Med 6: 621-631.
- Al-Muhsen S, Vazquez-Tello A, Jamhawi A, Al-Jahdali H, Bahammam A, et al. (2014) Association of the STAT6 rs324011 (C2892T) but not rs324015 (G2964A) with atopic asthma in a Saudi Arabian population. Hum Immunol 75:791-795.
- Godava M, Kopriva F, Bohmova J, Vodicka R, Dusek L, et al. (2012) Association of STAT6 and ADAM33 single nucleotide polymorphisms with asthma bronchiale and IgE level and its possible epigenetic background. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 156: 236-247.
- Pykalainen M, Kinos R, Valkonen S, Rydman P, Kilpeläinen M, et al. (2005) Association analysis of common variants of STAT6, GATA3, and STAT4 to asthma and high serum IgE phenotypes. J Allergy Clin Immunol 115: 80-87.
- Berenguer AG, Fernandes AT, Oliveira S, Rodrigues M, Ornelas P, et al. (2014) Genetic polymorphisms and asthma: findings from a case-control study in the Madeira island population. Biol Res 47: 40-51.
- Kim EG, Shin HJ, Lee CG, Park HY, Kim YK, et al. (2010)DNA methylation and not allelic variation regulates STAT6 expression in human T cells. Clin Exp Med 10: 143-152.
- Baek KJ, Cho JY, Rosenthal P, Crotty Alexander LE, Nizet V, et al. (2013) Hypoxia potentiates allergen induction of HIF-1 α, chemokines, airway in?ammation, TGF- β1, and airway remodeling in a mouse model. Clin Immunol 147: 27-37.
- Bao ZS, Hong L, Guan Y, Dong XW, Zheng HS, et al. (2011) Inhibition of airway inflammation, hyperresponsiveness and remodeling by soy isoflavone in a murine model of allergic asthma. Int Immunopharmacol 11: 899-906.
- Shi JP, Li XN, Zhang XY, Du B, Jiang WZ, et al. (2015) Gpr97 is dispensable for inflammation in ova-induced asthmatic mice. PLoS One 10: 398-403
- Wu YM, Xue ZW, Zhang LL, Gao NM, Du XM, et al. (2015) Comparable function of γ-tocopherols in asthma remission by affecting eotaxin and IL-4. Adv Clin Exp Med 2016; 4: 643-648.
- Nesi RT, Kennedy-Feitosa E, Lanzetti M, Ávila MB, Magalhães CB, et al. (2017) Inflammatory and oxidative stress markers in experimental allergic asthma. Inflammation 40: 1166-1176.
- Dunn RM, Wechsler ME (2015) Anti-interleukin therapy in asthma. Clin Pharmacol Ther 97: 55-65.
- Micheal S, Minhas K, Ishaque M, Ahmed F, Ahmed A (2013) IL-4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients. J Investig Allergol Clin Immunol 23: 107-111.
- Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC (2008) DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol 28: 139-146.
- Chen Y, Zhang Y, Xu M, Luan J, Piao S, et al. (2017) Catalpol alleviates ovalbumin-induced asthma in mice: Reduced eosinophil infiltration in the lung. Int Immunopharmacol 43: 140-146.
- Al-Quraishi GM (2013) Serum levels of total IgE, IL-12, IL-13 and IL-18 in children patients with asthma. Iraqi Journal of Pharmaceutical Sciences 22: 110-114.
- Noureldin M, Haroun M, Diab I, Elbanna S, Abdelwahab N (2014) Association of interleukin 13 +2044G/A polymorphism with bronchial asthma development, severity and immunoglobulin E levels in an Egyptian population. Diag Clin Med 4: 71-77.
- Alasandagutti ML, Ansari MS, Sagurthi SR, Valluri V, Gaddam S (2017) Role of IL-13 genetic variants in signalling of asthma. Inflammation 40: 566-577.
- Kupry?-Lipi?ska I, Moli?ska K, Kuna P (2016) The effect of omalizumab on eosinophilic inflammation of the respiratory tract in patients with allergic asthma. Pneumonol Alergol Pol 84: 232-243.
- De Vooght V, Smulders S, Haenen S, Belmans J, Opdenakker G, et al. (2013) Neutrophil and eosinophil granulocytes as key players in a mouse model of chemical-induced asthma. Toxicol Sci 131: 406-418.
- Nakagome K, Nagata M (2011) Pathogenesis of airway inflammation in bronchial asthma, Auris Nasus Larynx 38 : 555-563.
- Ma Y, Liu X, Wei Z, Wang X, Xu D, et al. (2014) The expression of a novel anti-inflammatory cytokine IL-35 and its possible significance in childhood asthma. Immunol Lett 162: 11-17.
- Malmström K, Lohi J, Sajantila A, Jahnsen FL, Kajosaari M, et al. (2017) Immunohistology and remodeling in fatal pediatric and adolescent asthma. Respir Res 18: 94-99.
- Motomura Y, Morita H, Moro K, Nakae S, Artis D, et al. (2014) Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity 40: 758-771.
- Cheng LE, Sullivan BM, Retana LE, Allen CD, Liang HE et al. (2015) IgE-activated basophils regulate eosinophil tissue entry by modulating endothelial function. J Exp Med 212: 513-524.
- Li J, Lin LH, Wang J, Peng X, Dai HR, et al. (2014) Interleukin-4 and interleukin 13 pathway genetics affect disease susceptibility, serum immunoglobulin E level and gene expression in asthma. Ann Allergy Asthma Immunol 113: 173-179.
- Oh CK, Geba GP, Molfino N (2010) Investigational therapeutics targeting the IL-4/IL-13/STAT-6 pathway for the treatment of asthma Eur Respir Rev 19: 46-54.



