Research Article - (2023) Volume 13, Issue 3
Received: 30-Aug-2023, Manuscript No. EJEBAU-23-17594; Editor assigned: 04-Sep-2023, Pre QC No. EJEBAU-23-17594 (PQ); Reviewed: 18-Sep-2023, QC No. EJEBAU-23-17594; Revised: 27-Sep-2023, Manuscript No. EJEBAU-23-17594 (R); Published: 13-Oct-2023, DOI: 10.36648/2248-9215.13.3.21
In the present study total 41 lines including a check, were evaluated in Randomized Block Design (RBD). For studying genetic diversity and Principle Component Analysis (PCA) using both uni and multi variant analysis. Based on D2 values, 41 genotypes were grouped into seven clusters. Cluster I with 4 genotypes, cluster II with 9 genotypes, cluster III with 5 genotypes, cluster IV with 2 and V with 5 genotypes cluster VI and VII with 8 genotypes in each. Non-hierarchical Euclidean cluster analysis using 11 quantitative characters revealed maximum diversity between cluster VI and VIII. The PF score creates a perfect base for the estimation of variability and for proper selection among complex characters. PCA-1 and PCA-2 combined together contributed with (6.04) of variation among seed yield characters. Based on cluster means, cluster distances and their performance C-1027, C-222, JGM-7, NBEG-47, IPC-10-134 were identified for inclusion in hybridization programme.
Chickpea; GCV; PCV; Genetic diversity; Cluster analysis; Principle component analysis
Chickpea also commonly known as bengal gram is an important cool season self pollinated legume. Comparatively with other legumes it tops in terms of nutrition and production with low cost of production which grows across the country. It is used famously used as dal. Chickpea is one of the oldest, cheapest sources of protein (approx. 21%) and it consists of aspartic acid followed by leucine and lysine. Chickpea is a king of pulses. Despite the fact having these many richer biochemical benefits in this drought tolerant legume still lacking in the productivity, to boost the production and productivity of chickpea, we need to develop improved varieties which are capable of tackling harsh conditions and resulting higher yields, for any improved breeding programme it is important to select robust and highly diverse parents, for that breeders need to evaluate the germplasm ruthlessly and figuring out better lines [1]. There are commonly two cultivated types of chickpeas which are desi (Cicer arietinum) and kabuli (Cicer kabulium) originated from south-east Asia (Afganisthan).
For studying this genetic diversity in and among the gene pool is possible through the both uni and multi variant statistical analysis such as cluster analysis and Principle Component Analysis (PCA). Broad knowledge of genetic diversity also aids in selecting superior parents over diverse genotype in hybridization programme. The present study is aimed to understand the genetic diversity among the germplasm with the help of wilks cluster analysis and principle component analysis to pick out possibly superior parents and exploit them further [2].
The experimental was carried out at the field experimentation centre of the department of genetics and plant breeding. The site is located at 25.28 N latitude, 81.54 E longitude it comes under sub-tropical and semi-arid climate. Lies 102 m above sea level. The average annual rainfall is 1042 mm. The experimental plot had sandy loam soil and normal fertility status. Sowing of 41 chickpea lines including check on 26th of November, 2019. Seed material was collected from the department of genetics and plant breeding were grown in Randomized Block Design (RBD) with three replications to check phenotypic and genotypic diversity [3]. Total eleven quantitative characters were chosen and taken into consideration for the estimation of genetic relationship among chickpea genotypes, the quantitative characters such as days to 50% flowering, days to 50% pod setting, plant height, number of primary branches per plant, number of pods per plant, number of seeds per plant, seed index (g), biological yield (g), harvest index (%) and grain yield per plant (g). The recorded data from the best 5 plants selected from the each plot further subjected to statistical analysis such as, Mahalanobis D2 statistic by Mahalanobis and Rao. The degree of divergence also plotted by using Principle Component Analysis (PCA) by using XLSTAT.
Genetic parameters of yield and yield components in chickpea lines the high Genotypic Coefficient of Variation (GCV) was found for number of pods per plant and economic yield (22%) followed by primary branches (21%) and biological yield (20%), while it was smaller in days to maturity (1%), days to 50% pod setting (4%) and days to 50% flowering (10%) [4]. Higher Phenotypic Coefficient of Variation (PCV) was also found in economic yield (25%) followed by number of pods per plant (23%) number of branches (22%) and biological yield (22%) while it was smaller in days to maturity (2%), day to 50% pod setting (4%) and days to 50% flowering (11%) and similar results were reported by Mirza et al.
Genetic parameters of yield and yield components in chickpea lines are shown in Figure 1. The high genotypic coefficient of variation was found for number of pods per plant and economic yield (22%) followed by primary branches (21%) and biological yield (20%), while it was smaller in days to maturity (1%), days to 50% pod setting (4%) and days to 50% flowering (10%) [5]. High phenotypic coefficient of variation was also found in economic yield (25%) followed by number of pods per plant (23%) number of branches (22%) and biological yield (22%) while it was smaller in days to maturity (2%), days to 50% pod setting (4%) and days to 50% flowering (11%).
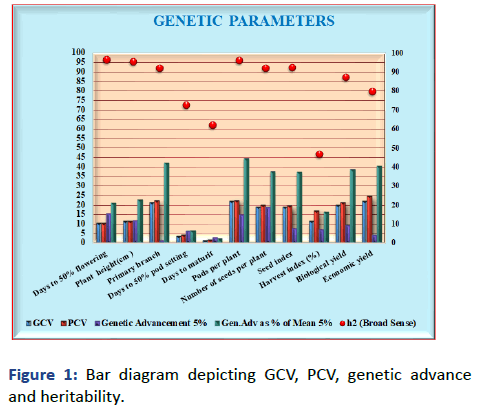
Figure 1: Bar diagram depicting GCV, PCV, genetic advance and heritability.
Genetic Divergence
Wilk’s criterion showed significant differences between the genotypes for the pooled effect of twelve characters studied. Hence, further analysis was done to calculate D2 values. The D2 values for all the possible pairs of comparison between 41 genotypes were calculated [6]. The calculated D2 value ranged from 3 to 130, the lowest value being for the character number of primary branches per plant and highest for days to maturity [7]. Based upon the observations of 11 characters, the Mahalanobis’s D2 statistics was computed for all possible pairs of 41 genotypes in order to assess the genetic diversity present among the genotypes under study (Tables 1 and 2).
| Sl. no | Cluster number | Number of genotypes | Genotypes included |
|---|---|---|---|
| 1 | I | 14 | NBEG-3, JG-24, IPC-06-77, ICC-1205, KSG-931, IPC-2K-25, IPC-05-24, IPC-6006, KPG-59, IPC-05-62, GNG-1581, GNG-1958, IPC-97-29, ICCY-10 |
| 2 | II | 14 | C-223, C-203, C-108, C-222, C-1028, C-130, C-1044, C-215, C-18125, C-213, UDAY, C-18126, C-210, C-207 |
| 3 | III | 3 | C-224, C-201, C-129 |
| 4 | IV | 1 | IPC-10-134 |
| 5 | V | 1 | NBEG-47 |
| 6 | VI | 3 | C-1027, ICC-16693, ICC-11019 |
| 7 | VII | 4 | ECSJ-627U, CSG-8962, JGM-7, C-1025 |
| 8 | VIII | 1 | IPC-04-52 |
Table 1: Distribution of genotypes into eight clusters.
| Cluster means: Tocher method | |||||||||||
| Days to 50% flowering | Plant height (cm) | Primary branches | Days to 50% pod setting | Days to maturity | Pods per plant | Number of seeds per plant | Seed index | Harvest index (%) | Biological yield | Economic yield | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cluster 1 | 82 | 54 | 4 | 98 | 126 | 30 | 50 | 25 | 47 | 27 | 12 |
| Cluster 2 | 67 | 54 | 3 | 99 | 129 | 36 | 47 | 18 | 43 | 20 | 9 |
| Cluster 3 | 66 | 59 | 3 | 92 | 130 | 47 | 55 | 17 | 40 | 29 | 12 |
| Cluster 4 | 81 | 48 | 3 | 99 | 128 | 33 | 50 | 24 | 40 | 25 | 10 |
| Cluster 5 | 83 | 47 | 3 | 92 | 126 | 32 | 46 | 24 | 41 | 28 | 11 |
| Cluster 6 | 67 | 42 | 3 | 99 | 129 | 28 | 59 | 16 | 42 | 18 | 8 |
| Cluster 7 | 78 | 41 | 3 | 97 | 126 | 31 | 49 | 24 | 42 | 29 | 12 |
| Cluster 8 | 81 | 64 | 4 | 101 | 128 | 31 | 69 | 22 | 47 | 29 | 14 |
Table 2: Cluster mean value for level quantitative traits.
Clustering Pattern of the Genotypes
The clustering pattern is obtained on the basis of magnitude D2 values. The 41 genotypes were grouped into seven clusters. Cluster I with 4 genotypes, cluster II with 9 genotypes, cluster III with 5 genotypes, cluster IV with 2 and V with 5 genotypes cluster VI and VII with 8 genotypes in each [8].
Cluster Means for Different Characters in Chickpea
The cluster mean for the eleven characters are presented in considerable inter-cluster variation was observed among the cluster means for the characters studied viz., days to 50% flowering, days to 50% pod setting, plant height (cm), number of branches, days to maturity, number of pods per plant, number of seeds per plant, seed index (g), biological yield (g), harvest index (%) and seed yield per plant (g). Cluster means for days to 50% flowering varied from 66 (III) to 83 (V). The lowest cluster mean for plant height was 48 (IV) and highest for cluster (III) 59. The cluster means for number of branches was ranged from 2.7 (II and VI) to 4.05 (I). Cluster means for days to 50% pod setting ranged between 92 (III and V) to 8 (VIII). The cluster means for days to maturity was ranged between 126 (I and V) to 130 (III). The cluster means for number of pods per plant was ranged between 30 (I) to 47 (III). The cluster means for number of seeds per plant varied from 46 (V) to 59 (VI). The cluster means for seed index was ranged between 16 (VI) to 25 (I). The cluster means for harvest index was ranged between 40 (III and IV) to 47 (I and VIII). The cluster means for biological yield 18 (VI) to 29 (III, VII and VIII). The cluster means for seed yield per plant was ranged between 8 (VI) to 12 (I, III and VII).
Percentage Contribution of Different Characters to Genetic Diversity
The utility of D2 analysis was enhanced by its application to estimate the relative contribution of the various plant characters to genetic divergence (Figure 2) [9]. The percent contribution of twelve characters studied was presented in Table 4. It was observed that, seed yield per plant (28.11), number of pods per plant (19.76), number of seeds per plant (17.20), biological yield (11.46), seed index (7.93), harvest index (6.95), days to 50% flowering (7.68), number of branches (4.02), days to 50% pod setting (1.22), plant height (0.98) and days to maturity (0.37) contributed least for genetic divergence.
Percentage Contribution of Different Characters to Genetic Diversity
The average intra and inter cluster D2 values and D values are presented in Figure 2. The inter cluster distance (D) range from 8.43 to 202.37. Highest inter cluster distance (D=202.37) was found between the cluster VI and VIII followed by the cluster VII and VIII (201.22), cluster VII and III (D=187.39). The minimum inter cluster distance (8.43) was between cluster V and IV. An examination of intra cluster divergence revealed that, cluster VII had maximum intra cluster distance. Similarly results was also projected by Jida and Alemu, Tamvar et al., Jakhar et al., Yadav et al., Lal et al., reported extensive genetic diversity in chickpea gene pool (Figures 3 and 4) [10].
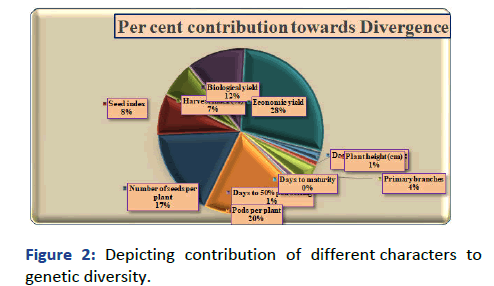
Figure 2: Depicting contribution of different characters to genetic diversity.
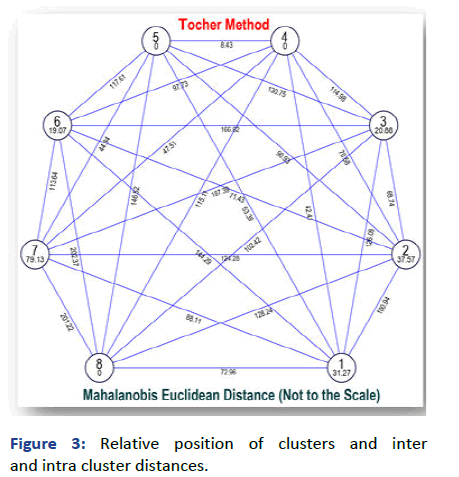
Figure 3: Relative position of clusters and inter and intra cluster distances.
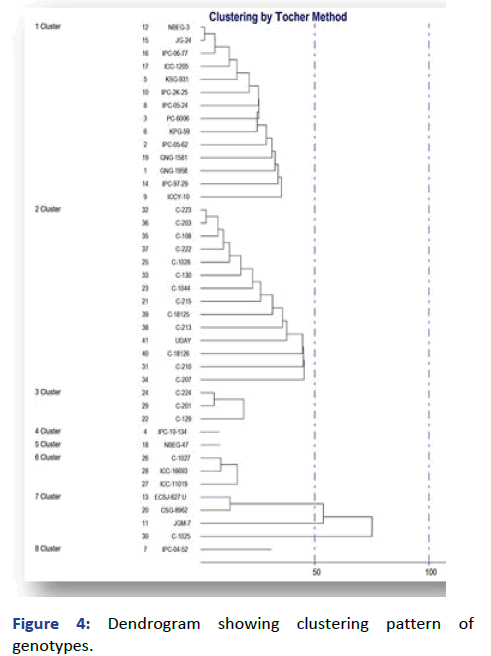
Figure 4: Dendrogram showing clustering pattern of genotypes.
Principle Component Analysis (PCA)
In these studies Principle Component Analysis (PCA) has been used to analyze the potent impact of different characteristics on grain yield (Tables 3 and 4). It is clearly noticed from the results, among the 4 PCA’s having eigen value about 8.57, out of which PCA-1 accounted for high proportion of variance (3.57%), followed by PCA-2 (2.47) and remaining two PCA’s showed (1.47%) and (1.07%) respectively (Figure 5).
| Canonical roots analysis (PCA) | |||||
| Sl.no | 1 Vector | 2 Vector | 3 Vector | 4 Vector | |
|---|---|---|---|---|---|
| Eigene value (Root) | 3.5705 | 2.4716 | 1.4734 | 1.0768 | |
| % Var. Exp. | 32.459 | 22.4695 | 13.3953 | 9.7893 | |
| Cum. Var. Exp. | 32.459 | 54.9292 | 68.3246 | 78.1139 | |
| 1 | Days to 50% flowering | 0.481 | 0.0163 | 0.1164 | 0.1011 |
| 2 | Plant height (cm) | -0.1221 | -0.5286 | 0.0424 | -0.2203 |
| 3 | Primary branches | 0.39825 | -0.2964 | 0.1501 | 0.2097 |
| 4 | Days to 50% pod setting | 0.26651 | -0.1403 | 0.2517 | -0.1626 |
| 5 | Days to maturity | -0.3634 | -0.3186 | 0.1885 | -0.1746 |
| 6 | Pods per plant | -0.0003 | -0.2816 | -0.6064 | -0.3076 |
| 7 | Number of seeds per plant | -0.0089 | -0.393 | 0.0104 | 0.6383 |
| 8 | Seed index | 0.49105 | -0.0073 | 0.0718 | -0.0852 |
| 9 | Harvest index (%) | 0.03327 | -0.1724 | -0.5653 | 0.4087 |
| 10 | Biological yield | 0.29164 | -0.3752 | -0.0778 | -0.3819 |
| 11 | Economic yield | 0.25406 | 0.3266 | -0.4056 | -0.1338 |
Table 3: PCA analysis of 11 quantitative characteristics.
| PCA I | PCA II | PCA III | ||
|---|---|---|---|---|
| Sl.no | Genotype | X Vector | Y Vector | Z Vector |
| 1 | GNG-1958 | 21.816 | -59.924 | 23.617 |
| 2 | IPC-05-62 | 22.651 | -58.921 | 24.173 |
| 3 | PC-6006 | 21.795 | -58.854 | 25.199 |
| 4 | IPC-10-134 | 20.117 | -54.234 | 24.308 |
| 5 | KSG-931 | 21.231 | -54.43 | 26.19 |
| 6 | KPG-59 | 23.27 | -58.218 | 24.976 |
| 7 | IPC-04-52 | 20.108 | -62.684 | 26.312 |
| 8 | IPC-05-24 | 23.05 | -58.036 | 24.409 |
| 9 | ICCY-10 | 23.1 | -59.476 | 23.968 |
| 10 | IPC-2K-25 | 21.035 | -55.921 | 24.579 |
| 11 | JGM-7 | 24.337 | -54.8 | 22.908 |
| 12 | NBEG-3 | 21.91 | -55.916 | 28.168 |
| 13 | ECSJ-627 U | 21.425 | -50.855 | 26.409 |
| 14 | IPC-97-29 | 22.192 | -59.921 | 27.189 |
| 15 | JG-24 | 21.779 | -55.198 | 27.83 |
| 16 | IPC-06-77 | 22.662 | -56.685 | 29.488 |
| 17 | ICC-1205 | 20.3 | -53.851 | 27.103 |
| 18 | NBEG-47 | 20.646 | -53.173 | 23.19 |
| 19 | GNG-1581 | 24.255 | -59.211 | 25.515 |
| 20 | CSG-8962 | 21.242 | -48.696 | 26.642 |
| 21 | C-215 | 12.871 | -54.746 | 25.223 |
| 22 | C-129 | 15.73 | -62.146 | 21.558 |
| 23 | C-1044 | 12.834 | -56.16 | 24.876 |
| 24 | C-224 | 13.93 | -60.217 | 21.79 |
| 25 | C-1028 | 15.952 | -54.356 | 24.895 |
| 26 | C-1027 | 14.329 | -51.418 | 25.963 |
| 27 | CC-11019 | 13.361 | -52.537 | 26.61 |
| 28 | CC-16693 | 15.467 | -51.818 | 25.788 |
| 29 | C-201 | 14.036 | -60.053 | 22.656 |
| 30 | C-1025 | 17.238 | -54.303 | 21.446 |
| 31 | C-210 | 14.917 | -57.505 | 22.125 |
| 32 | C-223 | 14.104 | -55.169 | 25.765 |
| 33 | C-130 | 14.708 | -57.485 | 26.518 |
| 34 | C-207 | 14.049 | -59.098 | 22.325 |
| 35 | C-108 | 14.796 | -54.09 | 25.592 |
| 36 | C-203 | 13.443 | -55.49 | 26.542 |
| 37 | C-222 | 14.593 | -56.968 | 25.916 |
| 38 | C-213 | 17.158 | -58.882 | 25.198 |
| 39 | C-18125 | 14.202 | -55.239 | 22.058 |
| 40 | C-18126 | 16.042 | -54.13 | 22.09 |
| 41 | UDAY | 17.06 | -53.341 | 24.06 |
Table 4: Total 3 PCA of 41 genotypes.
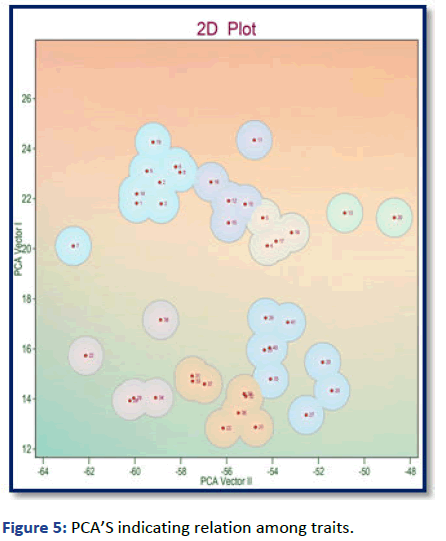
Figure 5: PCA’S indicating relation among traits.
The present studies analyzed by both cluster analysis and PCA resulted in the Principle Factors score (PF scores) for all the 41 genotypes. This PF score creates a ideal base for the estimation of variability and for proper selection among complex characters [11]. PCA-1 and PCA-2 combined together contributed (6.04) of variation among economic yield characters. The positive side of the PC-1 include characters such as seed yield per plant, number of primary branches, seed index, days to pod setting, harvest index and days to 50% lowering, genotypes which fall in this side are GNG-1958, IPC-0562, PC-6006, IPC-10-134, KSG-931 and KPG-59, while positive side of PC-3 includes days to 50% lowering, plant height, primary branches, days to 50% pod setting and days to maturity combined for them, genotypes which included are C-201, C-1025, C-210, C-223, C-130 and C-207, all the characters consists of higher genetic diversity (Figure 6) [12].
Figure 6: 3D plot for first 2 PCA’S.
All those characters can be further utilized to exploit heterosis from them and it also consists of broader genetic base. From the above results and the discussions, we can draw conclusions that the germplasm consisting of richer genetic diversity, which is upholding the genetic variability which is the important factors which need to consider by the breeder, the cluster and PCA analysis discloses the greater diversity which is already existed in the germplasm can be exploited further for hybridization programmes, to an extent traits such as seed yield, number of pods per plant and number of seeds per plant exhibiting greater diversity. Before initiating any breeding session we need to consider all the above stated factors such as, genetic variability and proper characteristics of parents should be contemplate in the irst place.
[Crossref] [Google Scholar] [PubMed]
Citation: Rao EG, Lavanya GR, Gowtham KV, Praneeth D, Hemalatha N (2023) Assessment of Genetic Diversity using Cluster and Principle Component Analysis in Chickpea (Cicer arietinum L.) Genotypes for Seed Yield Characters. Eur Exp Bio. 13:21.
Copyright: © 2023 Rao EG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction in any medium, provided the original author and source are credited.