Jacqueline C Alexander, Te-Wen Lo*, Jihye Lee, August P Miguez and Jason E Webb
Department of Biology, Ithaca College, Ithaca NY, USA
Corresponding Author:
Te-Wen Lo
Department of Biology, Ithaca College, Ithaca, NY, 14850, USA
Tel: (800) 429-4275
E-mail: twlo@ithaca.edu
Received date: July 24, 2018; Accepted date: August 04, 2018; Published date: August 06, 2018
Citation: Alexander JC, Lo TW, Lee J, Miguez AP, Webb JE (2018) A C. briggsae Genetic Suppressor Screen to Identify Dosage Compensation Pathway Components. Biochem Mol Biol J 4:17. doi: 10.21767/2471-8084.100066
Keywords
Genetic suppressor screen; C. briggsae; xol-1
Introduction
Proper development requires the precise regulation of gene expression. In addition to individual genes being required to be expressed at the right time and place during development, chromosome-wide gene regulation events, such as dosage compensation, are also required. Dosage compensation is a specialized mechanism of chromosome-wide gene regulation, by which the expression of X -linked genes are equalized between males, which have a single X chromosome and females, which have two. This process is essential in all heterogametic organisms and failure to properly equalize gene dose between heterogametic organisms often results in sexspecific lethality. Dosage compensation also provides us with an interesting situation in which to study evolution since numerous unrelated organisms have evolved different mechanisms to achieve this global regulation of chromosome activity. This process occurs differently in mammals (XX/XY), flies (XX/XY) and worms (XX/XO). Equalization of the X-linked gene products in mammalian males and females is achieved by shutting down one of the two X chromosomes in the somatic cells of female mammals [1]. In flies, the level of transcription of the single set of X-linked genes in the male (XY) is increased [2]. In worms, decreasing the level of transcription of both sets of each X-linked gene in hermaphrodites relative to males equalizes X-linked gene expression between the sexes [3].
The existence of different mechanisms indicates there is a high level of developmental variation for dosage compensation between different organisms. Each of these groups (mammals, flies, and worms) co-opted a different module of genes for dosage compensation, which resulted in three very different dosage compensation solutions mammals [1-3]. Therefore, to understand how this essential chromosome-wide gene regulation process has evolved, characterization and comparison of more closely related species is required. The two Caenorhabditis species, Caenorhabditis briggsae and Caenorhabditis elegans provide an ideal system for this analysis. C. elegans and C. briggsae separated ~15-30 million years ago, and their sequence divergence is about 0.3 substitutions per site, slightly greater than human and mouse [4].
In C. elegans, dosage compensation is achieved by sex-specific targeting of the DCC to the hermaphrodite X chromosome those results in a reduction of X chromosome transcript levels by one-half [3]. As a result, the gene dose from the two hermaphrodite X chromosomes and the single male X chromosome are equal. In C. elegans, the DCC is comprised of proteins that function in several essential cellular processes such as, dosage compensation, mitosis and meiosis [5]. For example, five DCC components (MIX-1, DPY-27, DPY-26, DPY-28, and CAPG-1) are homologous to subunits of condensin, a conserved protein complex that promotes the compaction, resolution, and segregation of chromosomes during mitosis and meiosis [6-10]. Additional DCC subunits, SDC-1, SDC-2, and SDC-3, confer sex-specificity to the dosage compensation process and recruit the DCC to the X chromosomes of hermaphrodites [11,12] resulting in a twofold reduction of X chromosome transcript levels [13,14] and hermaphrodite fates. This sex-specific targeting of the DCC is controlled by the expression of the developmental switch gene, xol-1 [15]. xol-1 is also responsible for regulating C. elegans sex determination [13]. High levels of XOL-1 result in a male fate (dosage compensation off), whereas low levels of XOL-1 results in a hermaphrodite fate (dosage compensation on) (Figure 1).
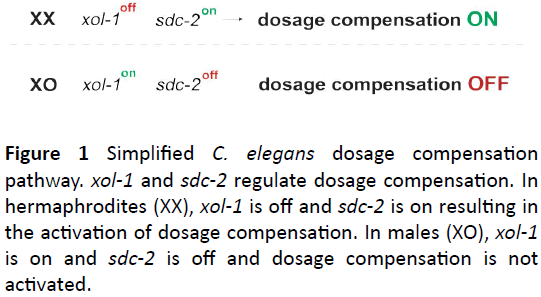
Figure 1: Simplified C. elegans dosage compensation pathway. xol-1 and sdc-2 regulate dosage compensation. In hermaphrodites (XX), xol-1 is off and sdc-2 is on resulting in the activation of dosage compensation. In males (XO), xol-1 is on and sdc-2 is off and dosage compensation is not activated.
While there is a wealth of knowledge about C. elegans dosage compensation and the role of xol-1 in regulation dosage compensation in C. elegans, far less is known about C. briggsae dosage compensation and the role of xol-1 in C. briggsae. Using a loss of function C. briggsae xol-1 deletion mutant (gift courtesy of B. J. Meyer), that results in male specific lethality we performed a genetic suppressor screen to identify components in the C. briggsae dosage compensation pathway including components of the C. briggsae dosage compensation complex itself.
Methods
Strains
• TY5006 C. briggsae xol-1(y430)
• RE921 C. brigssae him-8(v188)
• TWL005 C. briggsae him-8(v188); xol-1(y430)
C. briggsae him-8(v188); xol-1(y430) strain construction
Standard C. elegans genetic techniques [16] were used to build the C. briggsae him-8(v188); xol-1(y430) strain used in the suppressor screen. Briefly, Cbr-him-8(v188) males were crossed to Cbr-xol-1(y430) hermaphrodites. Cbr-him-8(v188)/+; +/xol-1(y430) heterozygous F1 progeny were allowed to produce self-progeny. F2 progeny were screened via PCR and Sanger sequencing to identify double homozygous C. briggsae him-8(v188); xol-1(y430) hermaphrodites.
C. briggsae xol-1 suppressor screen
C. briggsae him-8(v188); xol-1(y430) L4s were mutagenized using a traditional EMS mutagenesis screen [17]. L4 hermaphrodites were incubated in 47mM EMS for four hours. After EMS incubation, hermaphrodites were washed five times with M9 buffer and allowed to recover for one hour on an NGM plate with OP50 bacteria. After recovery, L4 hermaphrodites were cloned out (1/plate) and allowed to have progeny at 20°C until F1s reached the L4 stage and could be screened for the presence of males. F1 progeny of mutagenized hermaphrodites were also moved to new plates (3/plate) so that their F2 progeny could be screened for males.
Male rescue assay
Male rescue was determined by measuring the percentage of male and hermaphrodite progeny. Single L4 worms were cloned to individual NGM plates with OP50 and allowed to produce progeny at 20°C. Worms were moved to new NGM plates with OP50 every 24 hours until the worm was no longer laying embryos. Once the F1 progeny reached the adult staged they were scored as either hermaphrodites or males. For each individual L4 the percent male rescue=males/total progeny (males + hermaphrodites) produced by the individual. C. briggsae him-8, and him-8; xol-1 animals were used as controls. C. briggsae him-8 mutants produce 30% male progeny, and him-8; xol-1 mutants produce 0% male progeny.
Results
We utilized a suppressor screen (Figure 2) to identify downstream targets of C. briggsae xol-1. In both C. elegans and C. briggsae, the loss of XOL-1 results in a sex-specific male lethality. To facilitate this screen and remove the necessity of mating mutagenized C. briggsae xol-1 hermaphrodites to generate XO male progeny, we created a C. briggsae him-8; xol-1 double mutant. Mutations in him-8 result in a high incidence of male (XO) progeny from a self-fertile hermaphrodite.
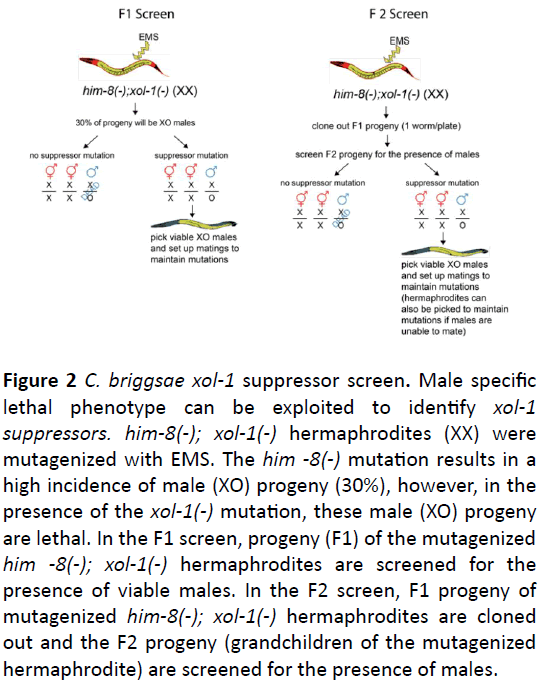
Figure 2: C. briggsae xol-1 suppressor screen. Male specific lethal phenotype can be exploited to identify xol-1 suppressors. him-8(-); xol-1(-) hermaphrodites (XX) were mutagenized with EMS. The him -8(-) mutation results in a high incidence of male (XO) progeny (30%), however, in the presence of the xol-1(-) mutation, these male (XO) progeny are lethal. In the F1 screen, progeny (F1) of the mutagenized him -8(-); xol-1(-) hermaphrodites are screened for the presence of viable males. In the F2 screen, F1 progeny of mutagenized him-8(-); xol-1(-) hermaphrodites are cloned out and the F2 progeny (grandchildren of the mutagenized hermaphrodite) are screened for the presence of males.
When combined with the xol-1 mutation, the C. briggsae him-8; xol-1 double mutant produces 30% XO male progeny that die as embryos due to xol-1 XO specific lethality [18]. We exploited this phenotype and performed a traditional EMS mutagenesis suppressor screen [17] to identify mutations that are able to suppress this xol-1 male lethal phenotype (Figure 2). We screened both F1 and F2 progeny of mutagenized worms. Screening both the F1 and F2 progeny allows for the ability to identify both dominant and recessive mutations. An additional advantage of screening F2 progeny is that it allowed for the identification of mutations that result in partial rescue of male lethality. For example, if the rescued male is unable to mate, the F2 screen allows for the isolation of the fertile hermaphrodite parent of a non-mating rescued male.
We screened 4447 haploid genomes and identified 9 suppressors that rescue the male lethal phenotype associated with loss of C. briggsae xol-1 function. We identified five suppressors (lot10, lot11, lot19, lot20, and lot23) from the F1 screen and four suppressors (lot21, lot22, lot24, and lot25) from the F2 screen. All suppressors were backcrossed (4X) and assayed for their ability to rescue the male lethal phenotype associated with loss of the C. briggsae xol-1 gene function (Figure 3). 10 L4s were cloned out for each suppressor strain and moved daily until worms stopped laying eggs. Once the F1 progeny reached adulthood, worms were scored as either hermaphrodite or male and percent male rescue was determined. Male rescue percentages ranged from 9% to 33% (Figure 3) demonstrating that there is variability between suppressors suggesting that our suppressors could represent alleles of more than one gene. The mutagenized strain, Cbrhim- 8(v188); xol-1(y430) produces 0% viable male progeny (Figure 3), therefore, the presence of any males indicates rescue. We used the Cbr-him-8(v188) strain as the positive control. This mutation results in approximately 30% male progeny (Figure 3), therefore, suppressors with 30% male progeny are considered to be fully rescued (Figure 3).
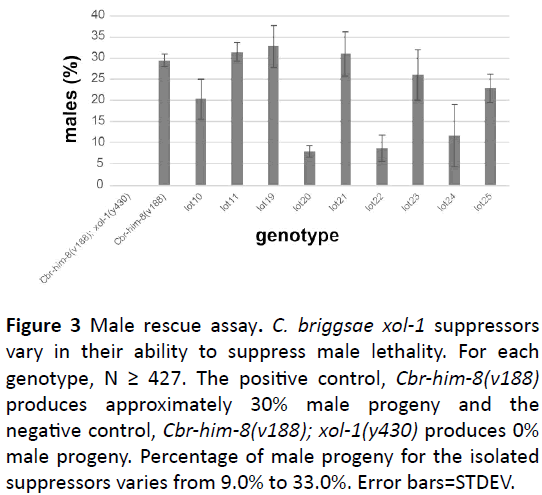
Figure 3: Male rescue assay. C. briggsae xol-1 suppressors vary in their ability to suppress male lethality. For each genotype, N ≥ 427. The positive control, Cbr-him-8(v188) produces approximately 30% male progeny and the negative control, Cbr-him-8(v188); xol-1(y430) produces 0% male progeny. Percentage of male progeny for the isolated suppressors varies from 9.0% to 33.0%. Error bars=STDEV.
Discussion and Conclusion
Using a traditional genetic suppressor screen (Figure 2), we isolated nine mutants that suppress the C. briggsae xol-1 male lethal phenotype. The nine suppressors vary in their ability to rescue the male lethal phenotype suggesting the isolation of different mutations (Figure 3). We will use SNP mapping and whole genome sequencing to identify the molecular lesions responsible for C. briggsae xol-1 suppression. We predict that suppressors will fall into three classes.
• Homologs of known C. elegans dosage compensation pathway components
• Novel components unique to C. briggsae that have homologs in C. elegans that have no known role in C. elegans dosage compensation
• Novel components unique to C. briggsae that have no known homologs in C. elegans.
All classes with further our understanding of the evolution of dosage compensation, however, the second and third classes (novel components) are particularly interesting. Novel components could either reveal divergence between mechanisms in C. elegans and C. briggsae or represent conserved components that have yet to be discovered in C. elegans allowing us to better understand dosage compensation in both species.
Acknowledgments
We would like to acknowledge B.J. Meyer and R. Ellis for sharing strains, E.S. Haag and L.E. Ryan for experimental assistance, and Ithaca College for funding.
References
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B (2003) Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36: 233-278.
- Meller VH, Kuroda MI (2002) Sex and the single chromosome. Adv Genet 46: 1-24.
- Meyer BJ, Casson LP (1986) Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47: 6: 871-881.
- Cutter AD (2008) Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol Biol Evol 25: 778-786.
- Csankovszki G, McDonel P, Meyer BJ (2004) Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 303: 1182-1185.
- Chuang PT, Albertson DG, Meyer BJ (1994) DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79: 3 459-474.
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ (1998) MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell 92: 265–277.
- Lieb JD, Capowski EE, Meneely P, Meyer BJ (1996) DPY-26, a Link Between Dosage Compensation and Meiotic Chromosome Segregation in the Nematode. Science 274: 1732-1736.
- Hirano T (2005) Condensins: Organizing and segregating the genome. Current Biol 15: R265-R275.
- Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, et al. (2008) Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev 22: 194-211.
- Davis TL, Meyer BJ (1997) SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome Development 124: 1019-1031.
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, et al. (1998) Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science 284: 1800-1804.
- Miller LM, Plenefisch JD, Casson LP, Meyer BJ (1988) xol-1: A gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55: 167-183.
- Meyer BJ (2000) Sex in the worm. Trends in Genetics 16: 247-253.
- Rhind NR, Miller LM, Kopczynski JB, Meyer BJ (1995) xo1-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell 80: 71-82.
- Fay D (2006) Genetic mapping and manipulation: Chapter 1-Introduction and basics. WormBook.
- Brenner S (1974) The Genetics of Caenorhabditis Elegans. Genetics 77: 71-94.
- Wei Q, Shen Y, Chen X, Shifman Y, Ellis RE (2014) Rapid Creation of Forward-Genetics Tools for C. briggsae Using TALENs: Lessons for Non model Organisms. Mol Biol Evol 31: 468-473.




